Abstract
By applying a novel software tool, information on 4080 UniGene clusters was retrieved from three adult human skeletal muscle cDNA libraries, which were selected for being neither normalized nor subtracted. Reconstruction of a transcriptional profile of the corresponding tissue was attempted by a computational approach, classifying each transcript according to its level of expression. About 25% of the transcripts accounted for about 80% of the detected transcriptional activity, whereas most genes showed a low level of expression. This in silico transcriptional profile was then compared with data obtained by a SAGE study. A fairly good agreement between the two methods was observed. About 400 genes, highly expressed in skeletal muscle or putatively skeletal muscle-specific, may represent the minimal set of genes needed to determine the tissue specificity. These genes could be used as a convenient reference to monitor major changes in the transcriptional profile of adult human skeletal muscle in response to different physiological or pathological conditions, thus providing a framework for designing DNA microarrays and initiating biological studies.
Preliminary analysis of collections of genes expressed in specific human tissues has been reported previously (Okubo et al. 1992; Itoh et al. 1994, 1998; Bernstein et al. 1996; Hwang et al. 1997; Maeda et al. 1997; Shimizu-Matsumoto et al. 1997; Bortoluzzi et al. 1998; Itoh et al. 1998). Expression profiles of human skeletal muscle involving <1000 genes have been produced in the past (Pietu et al. 1996; Lanfranchi et al. 1996). Although the most recent SAGE (serial analysis of gene expression) approach (Velculescu et al. 1995) promises to be more effective for such studies, to date it has been applied only to a limited number of tissues or cell types. Here, we report the application of a novel method for the retrieval and analysis of data from the UniGene database aimed at describing the transcriptional profiles of single human tissues. For this method, the expression level of each specific gene was quantitatively estimated under the assumption that the number of detected ESTs per gene is a function of its transcript frequency in the cell mRNAs population.
The UniGene database (Boguski and Schuler 1995) is a collection of ESTs corresponding to ∼90,000 putative human genes; it includes >5000 transcripts found in human skeletal muscle. Therefore, an in silico analysis of the transcription pattern of this tissue, based on UniGene data, is expected to produce a reasonable representation of its transcriptional profile. The present study reports on the analysis of 4080 transcripts found in human skeletal muscle, the estimated level of activity of the corresponding genes and their expression in different human tissues. A validation of this novel approach was attempted by a comparison with SAGE data on genes expressed in skeletal muscle (Welle et al. 1999).
RESULTS
This study is based on UniGene data from the three cDNA libraries most abundant in ESTs obtained from normal adult skeletal muscle without normalization or subtraction (Lib.24, Lib.272, and Lib.500). The automated retrieval and sorting of the corresponding UniGene entries, obtained by a software tool developed in our laboratory, produced a data set of 4080 individual transcripts. Only 1892 of them (46.4%) corresponded to mRNA gene sequences or to previously characterized human genes.
To estimate the size of the bias due to the possible correspondence of different UniGene clusters to the same gene, the UniGene entries considered by this study were classified according to the presence of 3′- and/or 5′-read ESTs in each UniGene cluster. Of 4080 UniGene entries, 3690 (90.4%) contained 3′- and 5′-read EST sequences, 52 (1.3%) contained only 5′-read ESTs, and 338 (8.3%) contained only 3′-read ESTs. Because at most, the 52 5′-read UniGene clusters could possibly refer to transcripts already identified by 3′-read clusters, the redundancy in our sample was <1.3%.
Our catalog of 4080 genes includes both skeletal muscle-specific genes and genes expressed in skeletal muscle as well as in other tissues. The level of expression of each entry of the catalog was quantitatively estimated by the number of the skeletal muscle ESTs corresponding to that given entry over the total number of skeletal muscle ESTs reported for all the entries included in the catalog. The basic assumption is that the number of detected ESTs per gene is a function of the transcript frequency in the population of mRNAs. The computation of the level of expression of each gene was automatically performed: The software analyzes the field ESTs SEQUENCES of each record, calculates the level of expression of the corresponding gene, and annotates the additional tissues in which the transcript was found.
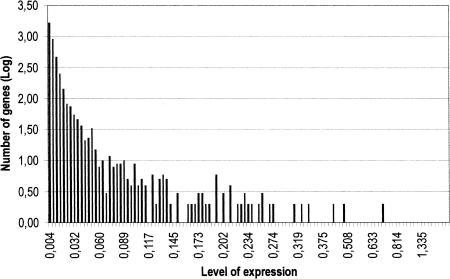
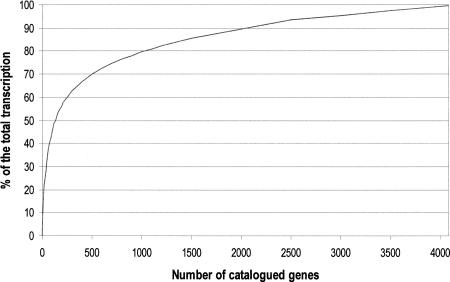
Figure 1 shows the frequency distribution of the number of genes per level of expression. The entries of the catalog were ranked in three different levels of expression: highly expressed genes (>0.0363% of the total detected transcription, corresponding to more than nine reported ESTs per entry), moderately expressed genes (from 0.0363% to 0.0121%; i.e., from nine to three ESTs per entry), and weakly expressed genes (<0.0121%, i.e., one or two ESTs per entry). The highly expressed genes accounted for 370 entries (9.1% of the total), the moderately expressed ones accounted for 1119 (27.4%) and the weakly expressed 2591 (63.5%). Figure 2 shows the cumulative percentage of the total transcriptional activity plotted against the number of genes. In the catalog, the genes were listed by decreasing level of expression.
Figure 1.
Frequency distribution of the number of genes per level of expression in the whole skeletal muscle data set (4080 genes).
Figure 2.
Cumulative percentage of the total transcriptional activity of 4080 genes expressed in human adult skeletal muscle, plotted against the number of genes reported in the catalog. The genes are listed in the same order as in the catalog.
The 100 most expressed genes do not include any of those expressed exclusively in adult skeletal muscle. The activity of skeletal muscle genes in other human tissues was evaluated by listing of the additional tissue(s) in which each entry of the catalog was reportedly expressed. Embryonic, fetal, infant, and tumor tissues were excluded from the analysis. Forty-seven entries (1.2% of the total) were found only in cDNA libraries obtained from human skeletal muscle; 27 of them were represented in UniGene by a single EST sequence. On the contrary, the large majority of skeletal muscle genes were found in at least one additional tissue and ∼89% in more than four additional tissues.
It is interesting to note that among the 34 entries corresponding to typically skeletal muscle genes (e.g., adult, skeletal, and sarcomeric form of myosin, actin, tropomyosin and telethonin, creatin kinase, aldolase, β enolase, titin, and myoglobin), 29 were determined to be highly expressed, and 22 occurred among the 100 most expressed genes. Grouping the 370 genes highly expressed in skeletal muscle and the 47 putatively skeletal muscle-specific genes, a set of 417 transcripts can be obtained, which may characterize the adult human skeletal muscle transcriptional profile.
Seventy-four entries corresponded to transcripts for ribosomal proteins, 21 to genes coding for translation factors (initiation or elongation), 2 to genes involved in DNA replication, and 14 to genes involved in DNA transcription (2 termination factors, 5 general transcription factors, 1 basic transcription factor, and 6 subunits of RNA polymerases I, II, or III). In total, this group accounted for 111 entries (3% of the total), presumably corresponding to sensu strictu housekeeping genes. They were found in up to 42 additional different tissues.
The results, obtained by the in silico reconstruction of the transcriptional profile of the skeletal muscle were compared with data from the Rochester Muscle Database (http://www.urmc.rochester.edu/smd/crc/Swindex.html), produced by the application of SAGE technology (Welle et al. 1999).
The Rochester SAGE catalog (July 1999) includes 12,207 unique species of muscle cDNA tags. We considered the expression data of 295 tags corresponding to genes highly expressed in skeletal muscle. The high expression level was deduced from the fact that the corresponding tags were detected >20 times over 53,875 independent sequences. All the selected tags corresponded to fully annotated genes. The gene product description and the GenBank ID were used for searching each gene in our catalog. The reciprocal correspondence among 120 genes belonging to both catalogs was established, and the level of expression of each gene was compared. The genes were classified into three categories according to the level of expression respectively estimated by the in silico or by the SAGE approach (class A, >2% of the total transcriptional activity detected in the tissue; class B, from 2% to 0.4%; class C, <0.4%). The proportions of genes falling to the three different categories in the two catalogs were compared by means of a χ2 test applied to a contingency table (Table 1), under the hypothesis of independence. The high value of χ2 (χ2 = 50.952, 4 d.f., P = 2.3 × 10−10) supports the hypothesis that the two distributions are significantly correlated. In addition, Fisher's exact test (Sokal and Rohlf 1998) was applied to test the null hypothesis of independence against the alternative hypothesis, without the χ2 approximation. The resulting P-value = 0 allowed us to discard the hypothesis of independence.
Table 1.
Contingency Table of Genes Falling in the Three Categories of Expression in the SAGE and in the In Silico Data Set
| In silico | Total | ||||
|---|---|---|---|---|---|
| A | B | C | |||
| A | 8 | 7 | 0 | 15 | |
| SAGE | B | 4 | 20 | 10 | 34 |
| C | 0 | 27 | 44 | 71 | |
| Total | 12 | 54 | 54 | 120 | |
Class A, >2%; class B, from 2% to 0.4%; class C, <0.4%. (First row) The number of genes falling in class A in the SAGE data set and falling in class A, B, or C in the in silico data set. (Second and third rows) The number of genes falling, respectively, in class B and C in the SAGE data set and in class A, B, or C in the in silico data set.
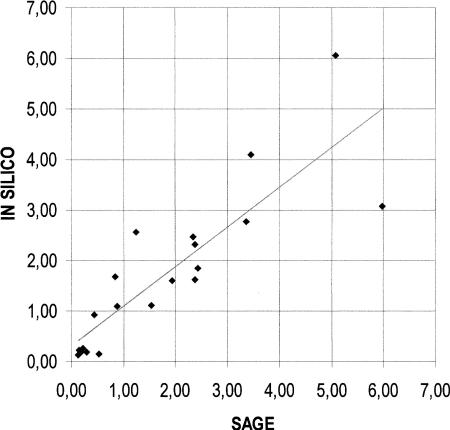
Twenty genes typical of skeletal muscle tissue were selected among the 120 reported in both catalogs (Table 2). Figure 3 shows the scatter plot of their levels of expression, independently calculated by in silico and SAGE methods. The linear correlation coefficient was determined to be 0.86.
Table 2.
List of 20 Genes, Typical of the Skeletal Muscle
| Gene | SAGE | In silico |
|---|---|---|
| α-actin, skeletal | 5, 97 | 3, 08 |
| creatine kinase M | 5, 08 | 6, 06 |
| myoglobin | 3, 46 | 4, 10 |
| myosin heavy chain 2a | 3, 36 | 2, 77 |
| myosin light chain 2 | 2, 44 | 1, 85 |
| desmin | 2, 38 | 1, 62 |
| fast skeletal troponin C | 2, 37 | 2, 32 |
| EST (slow twitch sk troponin 1) | 2, 34 | 2, 47 |
| β enolase | 1, 93 | 1, 60 |
| fast myosin alkali light chain 3 | 1, 53 | 1, 11 |
| titin | 1, 24 | 2, 56 |
| α-tropomyosin | 0, 87 | 1, 09 |
| telethonin | 0, 85 | 1, 67 |
| myosin alkali light chain 1f | 0, 53 | 0, 14 |
| tropomyosin | 0, 43 | 0, 93 |
| cytochrome c oxidase 5b | 0, 29 | 0, 20 |
| EST (myosin regulatory light chain 2) | 0, 23 | 0, 26 |
| cytochrome c oxidase 5a | 0, 19 | 0, 19 |
| creatine kinase, sarcomeric mitochondrial | 0, 15 | 0, 23 |
| α tubulin | 0, 13 | 0, 13 |
These genes were chosen from the group of 120 genes reported in the SAGE and the in silico catalog. For each gene, the estimated level of expression, with both methods, is provided.
Figure 3.
Scatter plot of the expression levels of a group of 20 typical skeletal muscle genes, reported in Table 2. On the x- and the y-axis are reported the level of expression estimated with the SAGE and with the in silico method, respectively. The best fitting trend line is shown. The correlation coefficient was determined to be 0.86.
DISCUSSION
The aim of this work was to develop an efficient method to mine the UniGene database to generate quantitative information on the human adult skeletal muscle transcriptome. UniGene is the largest database of expressed genes and is the least redundant among the present gene-indexing databases (Bouck et al. 1999). We estimated that the redundancy due to the existence of clusters containing only 5′- or 3′-read sequences is <1.3%. The fraction of UniGene clusters containing a very low number of sequences and/or not corresponding to known genes should be affected by a less negligible redundancy. Therefore, a correspondence between UniGene entries and genes may be legitimately assumed.
Among the human adult skeletal muscle libraries presently available in UniGene, we selected the three most abundant in ESTs, which have been neither normalized nor subtracted. Therefore, the 4080 individual transcripts resulting from the retrieval of UniGene records from these libraries should provide a fairly good sample, representative of genome expression in adult skeletal muscle. The total number of genes in the human genome is presently estimated to be between 80,000 and 140,000 (Dickson 1999). If it is assumed that <50% of these genes are active in a differentiated tissue, 40,000–70,000 genes are expected to be expressed in adult skeletal muscle. Our sample size, ∼8% of the presumed total, is much larger than the sample sizes currently used for statistical inference on unknown populations. On the other hand, the group of 4080 skeletal muscle genes considered here corresponds to 80% of the total number of genes expressed in muscle as reported so far in UniGene, fetal muscle and rhabdomyosarcoma included.
The computational approach in the analysis of expression profiles of cDNA libraries was introduced by Okubo et al. (1992) and later developed by Strausberg et al. (1997). This approach is unable to detect differences in gene expression due to post-transcriptional regulation processes, but the same limitation is shared by all of the present techniques attempting to estimate individual gene expression on a large scale. The software for the retrieval and the analysis of UniGene records used here for reconstructing the transcriptional profile of the skeletal muscle was used previously in our laboratory to construct genomic maps of genes expressed in specific human tissues (Bortoluzzi et al. 1998). The automated retrieval of the UniGene records and their sorting can be completed in an overnight session; on the contrary, the manual retrieval of the same information would take so long that it could hardly be performed before a new release of UniGene.
The use of the EST number per gene to quantify the gene expression is a principle adopted by several tools aiming to detect differences in gene expression activity, such as X-Profiler (http://www.ncbi.nlm.nih.gov/ncicgap/cgapxpsetup.cgi) and Digital Differential Display (http://www.ncbi.nlm.nih.gov/cgi-bin/UniGene/ddd?ORG=Hs; Strausberg et al. 1997). The present computational reconstruction of the transcriptional profile has the additional advantage of producing a general catalog (http://telethon.bio.unipd.it/GETProfiles/Skeletal_Muscle) in which each gene has a description, UniGene ID, and link for access to specific information (LocusLink, OMIM, SAGEMap). Moreover, this approach simplifies the statistical analysis and the comparison with other catalogs obtained from different tissues and with different methods.
The comparison of the present results with data obtained by SAGE on human adult skeletal muscle showed very good agreement between the two data sets. The comparison was limited to a relatively small number of entries because the Rochester Muscle Database has only a few annotated tags corresponding to nearly 300 genes for which the expression levels are available. The comparison was conducted in a very conservative way by use of only those genes for which the identity between the two catalogs was unequivocally established through the GenBank ID. Because the reliability of the SAGE approach is presently undisputed, the agreement between the two catalogs suggests that the in silico method may correctly reconstruct the transcriptional profile of a given tissue.
The picture emerging from the present study is that the transcriptional pattern of human adult skeletal muscle is characterized by a very low number of highly expressed genes, a tiny minority of them being tissue specific. Actually, 90% of the catalog is composed of genes moderately or weakly expressed. On the other hand, 417 genes account for about 50% of the total transcriptional activity of human adult skeletal muscle. Therefore, this group of genes may represent the minimal set needed to characterize the tissue.
Unexpectedly, the great majority of skeletal muscle genes were found in a number of tissues differing for structure, function, and embryological origin. Only one-tenth of genes were found expressed in less than five additional tissues and only 47 entries (1.2%) were found exclusively in skeletal muscle cDNA libraries. Possibly, this is due to the fact that all cells have a cytoskeleton and most exhibit some contractile properties. An additional explanation for the impressive sharing of transcripts among different tissues is that most of them share certain types of cells (e.g., fibroblasts, capillary endothelial cells, trapped blood cells). On top of that, the illegitimate transcription of several genes in a number of tissues may introduce an additional bias.
In our study the presence of skeletal muscle transcripts in additional tissues was recorded irrespective of the number of reported ESTs per transcript. It was impossible to circumvent the bias due to differences in number, size, and type of the original cDNA libraries.
EST sequencing, from which the in silico approach is derived, is intrinsically inadequate for identifying truly rare genes. On the other hand, if the sample size is sufficiently large, it may provide a fairly good quantitative estimation of the transcription of highly or moderately expressed genes (Audic and Claverie 1997). The same bias applies to the SAGE technique which, in addition, is affected by an underestimation of the very abundant transcripts. In the present study, the genes represented by one or two ESTs over 24,802 were considered as rare, whereas previous studies (Bernstein et al. 1996; Shimitsu-Matsumoto et al. 1997; Maeda et al. 1997) reported as rare those genes represented by a single EST over ∼1000. As the number of molecules of mRNA per nucleus is ∼150,000, a given transcript with a concentration of 10 molecules per nucleus would have about an 18% chance of remaining undetected in a sample of 25,000 ESTs. Therefore, the fact that a gene is not included in the catalog does not rule out the possibility that it is expressed in the tissue.
The approach proposed here represents a fast and almost inexpensive way to obtain fairly precise information on the transcriptional profile of different tissues. The limitation is the availability of sufficient data in UniGene, obtained from unnormalized or unsubtracted cDNA libraries. However, because the computational method is particularly effective in estimating the level of expression in highly expressed genes and because, presumably, every tissue is characterized by a relatively low number of highly expressed genes, this approach might be applied to situations where a relatively small number of transcripts are available.
This method appears to be a potentially powerful tool for high throughput electronic analysis (Bortoluzzi and Danieli 1998) of transcription patterns of the human genome. Because the information produced could be used as a reference for monitoring major changes in the transcriptional profile of a given human tissue in response to different physiological or pathological conditions, the in silico reconstruction of the transcriptional profile may prove particularly valuable for designing DNA microarrays and for initiating biological studies.
METHODS
UniGene is a collection of entries called UniGene clusters, each including all the ESTs corresponding to a putative gene. Each entry provides extensive information about EST sequences, genomic clones, complete mRNA (when available), gene name and description, function of the corresponding protein (if known), best SwissProt hits, mapping data, and tissue expression.
Three UniGene cDNA libraries obtained from normal adult skeletal muscle were selected for the study: Lib.24 Stratagene Human skeletal muscle cDNA library, cat. no. 936215 (2835 sequences), obtained from a sample of leg skeletal muscle of an adult female; Lib. 272 Stratagene muscle no. 937209 (5609 sequences), from the skeletal muscle of an adult male with malignant hyperthermia; Lib.500 HM3 (16,952 sequences), from a sample of skeletal muscle pectoralis maior of an adult female. These libraries have been neither normalized nor subtracted.
All the data presented in this paper were mined from the UniGene release no. 88 (July 1999), by a set of computer programs developed in our laboratory. The program RETRIEVE sends to the UniGene database server a continuous flow of queries pertaining to the selected libraries. The UniGene records, sent back by the server, are collected by the same program and merged in a single data set, removing the redundancy. Subsequently, the program SORT analyzes the different fields of each record and sorts out the entries according to a given criterion. In particular, for the present work, the field ESTs SEQUENCES was considered. For each entry, the number of ESTs obtained from skeletal muscle cDNA libraries was annotated and used for the computation of the level of expression in skeletal muscle. If additional ESTs, obtained from different tissues, were reported in the record, their presence was also automatically annotated by the program to produce information on the expression of the corresponding gene in other human tissues. The use of the software is offered free of charge to academics at the web site GETProfiles (http://telethon.bio.unipd.it/GETProfiles/index.html).
To analyze the transcriptional activity of human adult skeletal muscle, the number of ESTs corresponding to each UniGene entry found in the considered cDNA libraries was counted. The basic assumption is that the larger the number of skeletal muscle ESTs reported per entry, the more active the corresponding gene in the tissue. The number of ESTs per entry represents the best available estimate of the abundance of each individual transcript in the skeletal muscle mRNA population (Okubo et al. 1992; Itoh et al. 1994, 1998; Bernstein et al. 1996; Hwang et al. 1997; Maeda et al. 1997; Bortoluzzi et al. 1998).
The relative contribution of a given gene to skeletal muscle transcriptional activity is expressed by the ratio between the number of ESTs corresponding to that gene and the total number of ESTs corresponding to all the genes included in the data set. This ratio, expressed in percentage, will be referred as the level of expression of each gene.
The frequency distribution of genes according to their expression levels was analyzed by the descriptive statistics of the SPSS package (SPSS 8.0, SPSS Inc. Chicago USA). To assess the expression of skeletal muscle genes in other human tissues, each UniGene entry belonging to the data set was analyzed. The number and the type of tissues corresponding to the expression of each entry were recorded.
The human skeletal muscle expression profile obtained by the in silico reconstruction was compared with the SAGE data from the Rochester Muscle database (Welle et al. 1999). This database collects information on genes active in the human skeletal muscle and on their level of expression.
In July 1999, quantitative SAGE data regarding 295 annotated genes highly expressed in the skeletal muscle were available. The gene product description and the GenBank ID were used for searching each of these genes in the catalog produced by the present work, to establish the reciprocal correspondence between the genes in the two catalogs. For each gene, the number of ESTs in the in silico catalog and the number of TAG sequenced in the SAGE catalog were annotated and the percentage of the total was calculated over 24,802 muscle ESTs and 58,875 muscle tags, respectively. The values, representing the level of expression of the genes estimated with these two different methods, were normalized so that the sum of the percentage values, calculated over the 120 genes, equaled 100 in both catalogs.
According to their level of expression, reported respectively in each catalog, the genes were classified in three categories. The proportions of genes falling in the different categories in the two data sets were compared by a χ2 test, with Yates correction, applied to a 3 × 3 contingency table, with 4 d.f.. In addition, Fisher's Exact test was carried out, to test the null hypothesis of independence against the alternative hypothesis.
Acknowledgments
Stefania Bortoluzzi is a Ph.D. student of the Dottorato in Scienze Genetiche of the University of Ferrara. Fabio d'Alessi is recipient of a contract from TELETHON-Italy. The financial support of MURST is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
After this manuscript was accepted for publication, a paper appeared (Pietu et al., Genome Research 9: 1313–1320), where the expression profiling data of 910 genes expressed in human skeletal muscle were reported, with particular reference to 14 Genexpress IMAGE selected transcripts, 13 of which corresponded to UniGene clusters. Data are not directly comparable with those of the present paper. However, the analysis of the 13 transcripts suggests an agreement between the two catalogs. All of the 13 Genexpress IMAGE transcripts are included in the catalog reported in the present paper; 7 as highly expressed and 6 as moderately or weakly expressed.
Footnotes
E-MAIL danieli@bio.unipd.it; FAX 0039 049 8276209.
REFERENCES
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Bernstein SL, Borst DE, Neuder ME, Wong P. Characterization of a human fovea cDNA library and regional differential gene expression in the human retina. Genomics. 1996;32:301–308. doi: 10.1006/geno.1996.0123. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Schuler GD. ESTablishing a human transcript map. Nat Genet. 1995;10:369–371. doi: 10.1038/ng0895-369. [DOI] [PubMed] [Google Scholar]
- Bortoluzzi S, Danieli GA. Towards an in silico analysis of transcription patterns. Trends Genet. 1999;15:118–119. doi: 10.1016/s0168-9525(98)01682-5. [DOI] [PubMed] [Google Scholar]
- Bortoluzzi S, Rampoldi L, Simionati B, Zimbello R, Barbon A, d'Alessi F, Tiso N, Pallavicini A, Toppo S, Cannata N, Valle G, Lanfranchi G, Danieli GA. A comprehensive, high-resolution genomic transcript map of human skeletal muscle. Genome Res. 1998;8:817–825. doi: 10.1101/gr.8.8.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck J, Yu W, Gibbs R, Worley K. Comparison of gene indexing databases. Trends Genet. 1999;15:159–162. doi: 10.1016/s0168-9525(99)01709-6. [DOI] [PubMed] [Google Scholar]
- Dickson D. Gene estimate rises as US and UK discuss freedom of access. Nature. 1999;401:311. doi: 10.1038/43722. [DOI] [PubMed] [Google Scholar]
- Hwang DM, Dempsey AA, Wang RX, Rezvani M, Barrans JD, Dai KS, Wang HY, Ma H, Cukerman E, Liu YQ, Gu JR, Zhang JH, Tsui SK, Waye MM, Fung KP, Lee CY, Liew CC. A genome-based resource for molecular cardiovascular medicine: Toward a compendium of cardiovascular genes. Circulation. 1997;96:4146–4203. doi: 10.1161/01.cir.96.12.4146. [DOI] [PubMed] [Google Scholar]
- Itoh K, Okubo K, Yosii J, Yokouchi H, Matsubara K. An expression profile of active genes in human lung. DNA Res. 1994;1:279–287. doi: 10.1093/dnares/1.6.279. [DOI] [PubMed] [Google Scholar]
- Itoh K, Okubo K, Utiyama H, Hirano T, Yoshii J, Matsubara K. Expression profile of active genes in granulocytes. Blood. 1998;92:1432–1441. [PubMed] [Google Scholar]
- Lanfranchi G, Muraro T, Caldara F, Pacchioni B, Pallavicini A, Pandolfo D, Toppo S, Trevisan S, Scarso S, Valle G. Identification of 4370 expressed sequence tags from a 3′-end-specific cDNA library of human skeletal muscle by DNA sequencing and filter hybridization. Genome Res. 1996;6:35–42. doi: 10.1101/gr.6.1.35. [DOI] [PubMed] [Google Scholar]
- Maeda K, Okubo K, Shimomura I, Mizuno K, Matsuzawa Y, Matsubara K. Analysis of an expression profile of genes in the human adipose tissue. Gene. 1997;190:227–235. doi: 10.1016/s0378-1119(96)00730-5. [DOI] [PubMed] [Google Scholar]
- Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K. Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nat Genet. 1992;2:173–179. doi: 10.1038/ng1192-173. [DOI] [PubMed] [Google Scholar]
- Pietu G, Alibert O, Guichard V, Lamy B, Bois F, Leroy E, Mariage-Sampson R, Houlgatte R, Soularue P, Auffray C. Novel gene transcripts preferentially expressed in human muscles revealed by quantitative hybridization of a high density cDNA array. Genome Res. 1996;6:492–503. doi: 10.1101/gr.6.6.492. [DOI] [PubMed] [Google Scholar]
- Shimizu-Matsumoto A, Adachi W, Mizuno K, Inazawa J, Nishida K, Kinoshita S, Matsubara K, Okubo K. An expression profile of genes in human retina and isolation of a complementary DNA for a novel rod photoreceptor protein. Invest Ophthalmol Vis Sci. 1997;38:2576–2585. [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: The principle and practice of statistics in biological research. New York: W.H. Freeman and Co.; 1995. [Google Scholar]
- Strausberg RL, Dahl CA, Klausner RD. New opportunities for uncovering the molecular basis of cancer. Nat Genet. 1997;15:415–416. doi: 10.1038/ng0497supp-415. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Welle S, Bhatt K, Thornton CA. Inventory of high-abundance mRNAs in skeletal muscle of normal men. Genome Res. 1999;9:506–513. [PMC free article] [PubMed] [Google Scholar]