Abstract
Eye development in both invertebrates and vertebrates is regulated by a network of highly conserved transcription factors. However, it is not known what controls the expression of these factors to regulate early eye formation and whether transmembrane signaling events are involved. Here we establish a role for signaling via a member of the frizzled family of receptors in regulating early eye development. We show that overexpression of Xenopus frizzled 3 (Xfz3), a receptor expressed during normal eye development, functions cell autonomously to promote ectopic eye formation and can perturb endogenous eye development. Ectopic eyes obtained with Xfz3 overexpression have a laminar organization similar to that of endogenous eyes and contain differentiated retinal cell types. Ectopic eye formation is preceded by ectopic expression of transcription factors involved in early eye development, including Pax6, Rx, and Otx2. Conversely, targeted overexpression of a dominant-negative form of Xfz3 (Nxfz3), consisting of the soluble extracellular domain of the receptor, results in suppression of endogenous Pax6, Rx, and Otx2 expression and suppression of endogenous eye development. This effect can be rescued by coexpression of Xfz3. Finally, overexpression of Kermit, a protein that interacts with the C-terminal intracellular domain of Xfz3, also blocks endogenous eye development, suggesting that signaling through Xfz3 or a related receptor is required for normal eye development. In summary, we show that frizzled signaling is both necessary and sufficient to regulate eye development in Xenopus.
Vertebrate eye development begins with the formation of the eye field, a region within the anterior neural plate that gives rise to eyes (1). The eye field is demarcated by the expression of several regulatory genes, such as the homeodomain transcription factors Pax6, Rx, and Six3 (2–4). Pax6 in particular plays an essential role in defining the tissue that is fated to become eye, and appropriate expression is required for normal eye development. However, it is not yet known whether upstream signaling pathways control expression of these transcription factors and thus regulate eye development.
Proteins belonging to the wnt family of secreted ligands serve important patterning functions during development (5). Wnt proteins function by binding to seven-pass transmembrane receptors belonging to the frizzled family (6–9). Multiple wnt ligands and frizzled receptors are expressed during vertebrate nervous system development (10). For example, wnt signaling regulates formation of the neural crest (11) and anterior–posterior patterning of the central nervous system (12). One member of the frizzled family, Xenopus frizzled 3 (Xfz3), shows an expression pattern consistent with a role in regulating eye development. Xfz3 is expressed throughout development, but beginning at stage 12 expression increases and becomes restricted to the developing nervous system, with strong expression in the anterior neural plate in a region overlapping the early eye field (13). Xfz3 continues to be expressed in the developing optic vesicle as development proceeds (13). After neural tube closure Xfz3 is also expressed in the dorsal neural tube, where it regulates neural crest formation (M.A.D., J.-P. Saint-Jeannet and P.S.K., unpublished observations).
Several wnt ligands are also expressed during eye development, although none have been specifically implicated at the early stages of this process. Wnts activate frizzled receptors by binding to the cysteine-rich extracellular domain of the receptor (7). Frizzled activation can lead to signaling through either a canonical pathway involving β-catenin or noncanonical pathways that regulate planar cell polarity in Drosophila and possibly vertebrates (14–16), as well as calcium mobilization and protein kinase C activation in Xenopus and zebrafish (17). Additional novel proteins have been identified that may couple frizzled receptors to these downstream signaling pathways. In the case of Xfz3, a protein called Kermit has been identified that specifically interacts with the cytoplasmic C-terminal domain of the receptor (C.T., M.A.D., J.-P. Saint-Jeannet, and P.S.K., unpublished observations). Kermit is an intracellular PDZ-domain protein that is expressed in the nervous system and developing eye tissue in a spatial and temporal pattern very similar to that of Xfz3 and appears to be required to transduce signaling through Xfz3.
Here we implicate frizzled signaling in the regulation of eye formation. We demonstrate that overexpression of Xfz3 is sufficient to promote complete ectopic eye development and to promote ectopic expression of the eye regulatory genes Xpax6, Xrx, and Xotx2. In addition, overexpression of a soluble inhibitory form of Xfz3 blocks endogenous eye development and blocks endogenous expression of the same eye regulatory genes. Finally, inhibition of signaling through Xfz3 by overexpression of Kermit, a protein that binds to the C-terminal intracellular region of Xfz3, also blocks endogenous eye development. In summary, these findings demonstrate a requirement for wnt/frizzled signaling in regulating vertebrate eye development.
Materials and Methods
Constructs and Microinjection of RNA.
Xfz3 (13) was independently isolated in a screen to identify Xenopus frizzled homologs (18). The ORF was subcloned into pCS2+, with EcoRI and XhoI sites added by PCR. To generate Nxfz3, the region encoding amino acids 1–202 was subcloned into pCS2+MT with the use of BamHI and ClaI sites added by PCR (M.A.D. and P.S.K., unpublished observations). Capped RNA was synthesized in vitro by SP6 transcription from pCS2+-Xfz3, pCS2MT-Nxfz3, pCS2+Kermit, or pCS2+-nuclear β-galactosidase (nβgal) template DNA with a Message Machine kit from Ambion (Austin, TX). For eight-cell stage injections, RNA was injected in a volume of 1 nl into one of the dorsal animal blastomeres in the following amounts (unless indicated otherwise): Xfz3 (2 ng), Nxfz3 (2 ng), and nβgal (0.1 ng). For the rescue experiments 2 ng of Xfz3 RNA was coinjected with 2 ng of Nxfz3 RNA. Kermit RNA was injected at the four-cell stage into the animal pole region of a dorsal blastomere (0.25–2 ng). The embryos were staged according to the method of Nieuwkoop and Faber (19) and fixed in MEMFA (0.1 M Mops, pH 7.4/2 mM EGTA/1 mM MgSO4/3.7% formaldehyde) (20) at the stages indicated in the text. 5-Bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) staining was performed on embryos injected with βgalactosidase RNA as previously described (21).
Sectioning and Immunohistochemistry.
For immunohistochemistry, embryos were fixed at stage 42–45 and sectioned on a cryostat at a thickness of 14 μm. Immunohistochemistry was performed with the use of a mouse anti-rhodopsin antibody (22) (1:100) and rabbit anti-Pax6 antibody (23) (1:250), by methods described previously (21). The primary antibodies were detected with an Alexa Fluor 488 goat anti-mouse IgG antibody or an Alexa Fluor 568 goat anti-rabbit IgG antibody (Molecular Probes; 1:300). The sections were stained with Hoechst dye (30 μM in PBS) to visualize the nuclei and coverslipped with the use of Fluoromount-G mounting media (Southern Biotechnology Associates). Fluorescent images were acquired on a Nikon E800 microscope with a Xillix PMI camera and openlab software. For sections that were not immunostained, embryos were fixed at stage 42–45, embedded in paraffin, sectioned at a thickness of 14 μm, and then dewaxed and coverslipped with the use of Pro-Texx mounting medium (Baxter Diagnostics, McGaw Park, IL).
In Situ Hybridization.
Digoxigenin-labeled antisense RNA probes were generated for XPax6 (24), Xrx (3), and Xotx2(25) with the use of a Maxiscript RNA synthesis kit (Ambion) and digoxigenin-labeled UTP (Roche Molecular Biochemicals). Whole-mount in situ hybridization was performed on albino Xenopus embryos as previously described (20), except that either BM purple (Roche Molecular Biochemicals) or magenta-phos (Biosynth, Basel) was used as the substrate for the alkaline phosphatase reaction.
Results
Overexpression of Xfz3 Promotes Ectopic Eye Formation and Perturbs Endogenous Eye Development.
In the developing Xenopus nervous system Xfz3 is expressed in the anterior neural plate overlapping the region fated to give rise to eyes (13). To determine whether Xfz3 can influence eye development, we injected RNA encoding Xfz3 along with RNA for the lineage tracer β-galactosidase into a dorsal animal (D1) blastomere of an eight-cell embryo, which makes a major contribution to head ectodermal and mesodermal structures (26). When embryos were scored at stage 41–45, in multiple independent experiments we observed several eye-related phenotypes. In a representative experiment 11% (9/79) of the embryos had two endogenous eyes along with a third ectopic eye that was roughly spherical, surrounded with dense pigment and associated with lens-like tissue (Fig. 1 a and b). The ectopic eyes appeared on the dorsal aspect of the embryo at or just lateral to the midline and were always positioned rostral to the spinal cord and caudal to the endogenous eyes. Twenty-five percent (20/79) of the embryos showed defects in the endogenous eye on the injected side of the embryo. These defects included eyes in which the retinal pigment epithelium streamed toward the midline (Fig. 1c) and eyes that were placed more medially (Fig. 1 d and e). In 15% (12/79) of the embryos we observed isolated dense patches of pigment that resembled ectopic retinal pigment epithelium. These patches of ectopic pigment were fairly unrestricted in their location on the embryo, and the positions in which they appeared depended on the blastomere injected. Twenty-four percent (19/79) of the embryos had no eye phenotypes, but showed a shortened anterior–posterior axis and were often also bent dorsally, which is similar to what has been reported for injection of Xfz3 RNA at the two-cell stage (13), whereas 24% (19/79) of the embryos appeared morphologically normal. We observed none of the above eye phenotypes when Xfz3 RNA was injected at the two-cell stage. In addition, no ectopic eyes or defects in the endogenous eyes were observed when Xfz3 RNA was injected into ventral animal blastomeres (0/84 embryos), which make a much smaller contribution to anterior neural structures (26). However, 6% (5/84) of these embryos showed patches of ectopic pigment. Finally, no effects were observed when equivalent levels of RNA encoding β-galactosidase or green fluorescent protein were injected (data not shown). Notably, the eye phenotypes obtained with Xfz3 overexpression were remarkably similar in nature and frequency to those observed with Pax6 overexpression in Xenopus, including the formation of complete ectopic eyes and perturbation of endogenous eye formation (27). Overall, these findings suggest that Xfz3 is activating a specific signaling pathway that is sufficient to promote eye development.
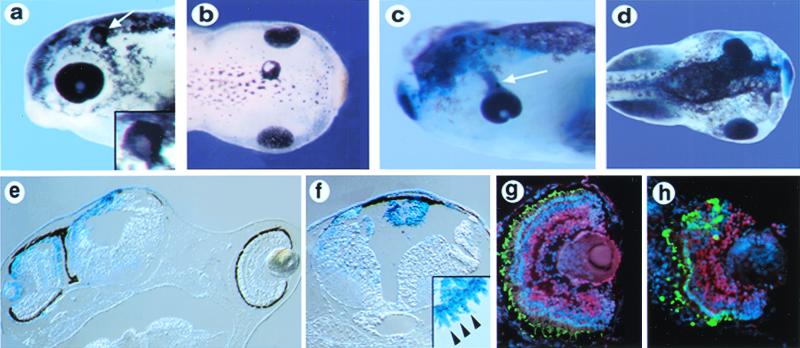
Figure 1.
Overexpression of Xfz3 promotes ectopic eye formation and causes proximal eye defects. (a) Stage 42 embryo showing an ectopic eye in the dorsal head region (white arrow). (Inset) A higher magnification view of the ectopic eye. (b) Stage 45 embryo with an ectopic eye in the dorsal midline of the head. (c) Stage 42 embryo showing streaming of the retinal pigment epithelium from the endogenous eye toward the midline. (d) Stage 45 embryo with abnormally positioned eye on the injected side, marked with β-galactosidase (blue). (e) Section through an embryo similar to the one shown in d, showing the eye adjacent to the neural tube on the injected side (Left). (f) Section through a stage 45 embryo, showing an ectopic eye in the roof of the fourth ventricle that is fully labeled with the tracer β-galactosidase (blue). (Inset) A high-magnification view from an adjacent section, showing a row of morphologically distinct photoreceptor outer segments in the ectopic eye (arrowheads). (g and h) Immunolabeling of a section through a normal eye (g) and ectopic eye (h) with anti-Pax6 antibodies (red) and anti-rhodopsin antibodies (green). Nuclei are labeled with Hoechst (blue). The ectopic eye in h shows an arrangement of retinal layers similar to that in the normal eye in g.
Ectopic eyes induced by Xfz3 overexpression were frequently located in the roof of the fourth ventricle, with photoreceptors invariably located in the outermost layer and oriented toward the lumen of the ventricle (Fig. 1f). The photoreceptors were always organized into a defined layer, had morphologically distinct outer segments (Fig. 1f; see Inset), and were labeled with anti-rhodopsin antibodies (data not shown; 5/5 embryos). Inclusion of RNA encoding the lineage tracer β-galactosidase in the injections showed that the ectopic eyes were derived exclusively from tissue expressing Xfz3, demonstrating that Xfz3 acted cell autonomously to promote eye formation (Fig. 1f). The laminar organization of the ectopic eyes was similar to that of endogenous eyes (compare Fig. 1 g and h). The retina was surrounded by retinal pigment epithelium, and adjacent to this were rhodopsin-positive photoreceptors with outer segments. Pax6-positive cells located more internally indicated the presence of retinal ganglion cells and amacrine cells (Fig. 1h). In addition, morphologically distinct lens tissue was often associated with the ectopic eye (Fig. 1h). These findings demonstrate that Xfz3 can function cell autonomously to promote the formation of fully differentiated ectopic eyes.
Overexpression of Xfz3 Activates Ectopic Expression of Xpax6, Xrx, and Xotx2.
The ability of Xfz3 to promote eye development suggested that it could be acting on the network of transcription factors that regulates the earliest stages of eye development. We therefore examined the expression of the homeodomain transcription factors Xpax6 and Xrx, which are expressed in the early eye field in vertebrates and are required for normal eye development (2, 3). We also examined the expression of Xotx2, which begins expression earlier and is critical for normal development of structures derived from the anterior neural plate (28, 29). Embryos were injected at the eight-cell stage with a mixture of RNA for Xfz3 and β-galactosidase, which was used to mark the region of the embryo derived from the injected blastomere. The embryos were collected for whole-mount in situ hybridization analysis at stage 14, to determine whether Xfz3 influences the expression of these genes in the early eye field, and at stage 28 to determine whether the later pattern of expression of these genes was affected. At stage 14, overexpression of Xfz3 resulted in expanded or ectopic Xotx2 expression in 13% of embryos (3/23; Fig. 2f) and weakly expanded Xrx expression in 25% of embryos (4/16; Fig. 2d), but caused no change in Xpax6 expression (17/17 embryos; Fig. 2b). At stage 28, 12% of embryos (2/17) showed ectopic Xotx2 expression (Fig. 2l), 21% of embryos (6/29) showed ectopic Xrx expression (Fig. 2j), and 11% of embryos (3/28) showed ectopic Xpax6 expression (Fig. 2h). The frequency of ectopic gene activation roughly paralleled the frequency of ectopic eye formation observed with Xfz3 overexpression. In addition, the ectopic gene expression at stage 28 was often located in the dorsal part of the embryo in the general location where ectopic eyes frequently formed. At stage 28, when β-galactosidase expression directly overlay the endogenous eye, we observed some reduction of Xrx expression (7/29 embryos) and Xotx2 expression (5/17 embryos) on the injected side (data not shown), consistent with Xfz3 overexpression causing proximal eye defects (e.g., misplaced eyes). In summary, these findings demonstrate that overexpression of Xfz3 is sufficient to cause ectopic expression of eye regulatory genes. This effect was most apparent at later stages of development, consistent with either later availability of wnt ligand to activate the frizzled receptor or a delay in the competence of the responding tissue.
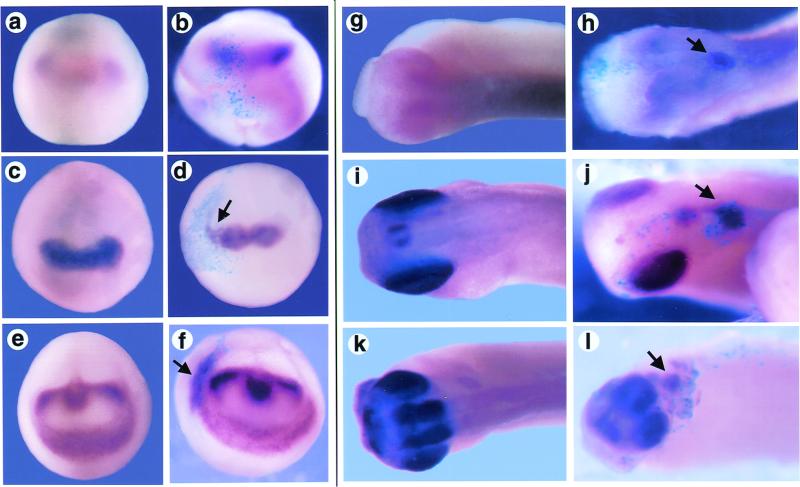
Figure 2.
Overexpression of Xfz3 promotes ectopic expression of genes regulating eye development. A whole-mount in situ hybridization analysis was made of stage 14 embryos (a–f) that were either uninjected (a, c, and e) or Xfz3 injected (b, d, and f) and then labeled for Xpax6 (a and b), Xrx (c and d), or Xotx2 expression (e and f). Arrows indicate modest expansion of Xrx or Xotx2 expression (d and f). Stage 28 embryos (g–l) are either uninjected (g, i, and k) or Xfz3 injected (h, j, and l) and then labeled for Xpax6 (g and h), Xrx (i and j), or Xotx2 expression (k and l). Arrows indicate ectopic expression in injected embryos (h, j, and l). β-Galactosidase (sky blue) marks the regions of the embryo derived from the injected blastomere.
Overexpression of an Inhibitory Form of Xfz3 Blocks Endogenous Expression of Xpax6, Xrx, and Xotx2 and Blocks Normal Eye Development.
The ability of Xfz3 overexpression to induce ectopic expression of eye regulatory genes suggested that Xfz3 could play a role in regulating their expression during normal development. To determine whether Xfz3 function was required for the expression of Xpax6, Xrx, and Xotx2 we overexpressed an inhibitory form of Xfz3 (Nxfz3) by RNA injection into a dorsal blastomere of eight-cell embryos. Nxfz3 consists of the extracellular cysteine-rich domain of the Xfz3 protein, a region in frizzled receptors that has been implicated in the binding of wnt ligand (6). Nxfz3 can block interaction of wnt ligands with the endogenous Xfz3 receptor and has been shown to interfere with signaling through Xfz3 (M.A.D., J.-P. Saint-Jeannet, and P.S.K., unpublished observations). After injection of RNA for Nxfz3 along with RNA encoding βgalactosidase, embryos were grown until stage 14 (open neural plate stage) or stage 18–20 (late neurula) and analyzed by whole-mount in situ hybridization for expression of Xpax6, Xrx, or Xotx2. At stage 18–20, in embryos where β-galactosidase signal was localized in the region of the developing eye field (anterior neural tube), there was reduced Xpax6 expression or Xpax6 expression was absent on the injected side in 38% (9/24) of the embryos (Fig. 3b). Similarly, Xrx expression and that of Xotx2 were reduced or absent in 48% (15/31) and 33% (7/21) of the embryos, respectively (Fig. 3 d and f). In embryos where β-galactosidase expression was not localized to the developing eye field, expression of Xpax6, Xrx, and Xotx2 was normal. Inhibition of gene expression was apparent from the onset of eye field formation, inasmuch as at stage 14 expression of Xpax6 (9/18 embryos), Xrx (10/25 embryos), and Xotx2 (6/23 embryos) was suppressed on the injected side (data not shown). These findings demonstrate that Nxfz3 is able to interfere with the expression of key regulatory genes controlling eye development and suggest that normal frizzled signaling is required for early expression of these genes.
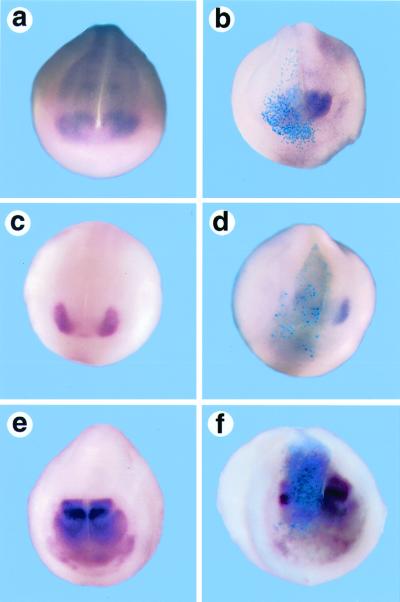
Figure 3.
Overexpression of Nxfz3 suppresses expression of genes regulating eye development. A whole-mount in situ hybridization analysis was made of stage 18–20 embryos that are either uninjected (a, c, and e) or Nxfz3 injected (b, d, and f) and then labeled for Xpax6 (a and b), Xrx (c and d), or Xotx2 expression (e and f). β-Galactosidase (sky blue) marks the regions of the embryo derived from the injected blastomere.
Because loss of Pax6, Rx, or Otx2 expression results in disrupted eye development in mammals (2, 3, 28, 29), we tested whether overexpression of Nxfz3 in Xenopus embryos would interfere with normal eye development. Sixty-three percent (62/98) of the embryos analyzed at stage 45 that were injected with Nxfz3 RNA at the eight-cell stage showed reduced or malformed eyes (Fig. 4b), and 7% (7/98) were missing eyes entirely (Fig. 4c), with only residual disorganized pigment in their place (Fig. 4d). There were no general effects on anterior development, inasmuch as the olfactory pits and cement gland on the injected side were intact (data not shown). In addition, the neural tube in Nxfz3-injected embryos looked morphologically normal (Fig. 4d). To demonstrate that the effect of Nxfz3 on endogenous eye development was due to interference with Xfz3 function, we rescued the effects of Nxfz3 by coexpressing full-length Xfz3 and observed that 88% (43/49) of the embryos were normal, with only 12% (6/49 embryos) showing reduced or missing eyes. No effects on eye development were observed when the tracer β-galactosidase was not localized to the head region. These findings demonstrate that normal frizzled signaling, potentially through Xfz3, is required for endogenous eye development.
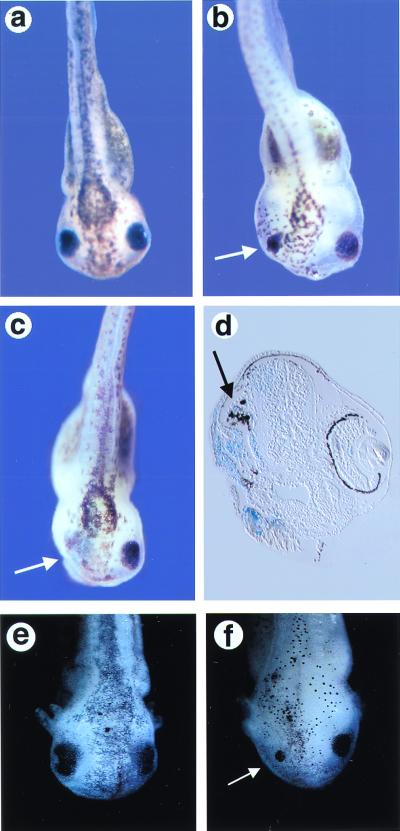
Figure 4.
Overexpression of Nxfz3 or Kermit inhibits endogenous eye formation. (a) Uninjected embryo at stage 41. (b and c) Nxfz3-injected embryos at stage 41 showing a reduced (b) or absent eye (c) on the injected side (white arrow). (d) Section through an embryo with a missing eye, showing the loss of normal eye structure on the injected side with only residual disorganized pigment remaining (black arrow). The tissue derived from the injected blastomere is marked with β-galactosidase (sky blue). (e) Uninjected embryo at stage 43. (f) Kermit-injected embryo at stage 43 showing a reduced eye on the injected side (arrow).
Overexpression of Kermit, a Xfz3-Interacting Protein, Inhibits Endogenous Eye Development.
To further test whether signaling through Xfz3 is required for endogenous eye development we have assessed whether Kermit, a component of the signaling pathway immediately downstream of Xfz3, can modulate this process. Overexpression of Kermit inhibits signaling through Xfz3, likely by swamping out interactions between the receptor and downstream signaling components. Kermit was overexpressed by injecting RNA into the animal pole region of a dorsal blastomere of four-cell stage embryos along with RNA encoding β-galactosidase. The majority of embryos analyzed at stage 40–45 that were injected with Kermit RNA at the eight-cell stage showed reduced eyes (58%, 193/331 embryos; Fig. 4f), and some were missing eyes entirely (7%, 24/331 embryos), whereas only 1/67 uninjected embryos had a reduced eye. These findings provide independent evidence that perturbing wnt/fz signaling can disrupt normal eye development.
Discussion
We show that wnt/fz signaling can regulate normal eye development and can also initiate ectopic eye formation. Overexpression of Xfz3 is sufficient to initiate expression of eye-specific genes and promote the formation of differentiated retinal tissue. We also provide evidence that frizzled signaling is required for endogenous eye development. Overexpression of a soluble inhibitory form of Xfz3 blocks endogenous eye development and blocks endogenous expression of Xpax6, Xrx, and Xotx2. In addition, inhibition of signaling through Xfz3 by overexpression of Kermit, a protein that binds specifically to the C-terminal intracellular region of Xfz3, also blocks endogenous eye development. Our data provide evidence of an extracellular signaling pathway that regulates normal and ectopic eye formation.
Our data suggest that Xfz3 is the frizzled receptor involved in eye formation during normal development. Xfz3 is expressed in the anterior neural plate in the region of the early eye field in concert with other early eye markers (13). Other members of the frizzled family such as Xenopus frizzled 2 (Xfz2) and zebrafish frizzled 9 (Zfz9) can also promote ectopic eye formation at frequencies similar to that of Xfz3 (J.T.R., M.S.R., and M.L.V., unpublished observations). However, Fz9 is not expressed in the developing eye in any species examined so far (T. Van Raay, J.T.R., M.S.R., and M.L.V., unpublished observations), making it an unlikely candidate, and Xfz2 is expressed in the eye at later developmental stages after eye formation has been initiated. The ability of these receptors to promote ectopic eye formation is likely due to overlap between frizzled receptors with respect to ligand binding specificity and the activation of downstream signaling pathways.
Overexpression of Xfz3 promoted the formation of complete ectopic eyes. To date, only a few proteins, all homeobox-containing transcription factors, have been shown to be sufficient to induce ectopic eye development in vertebrate embryos. In Xenopus and medaka, these include Xpax6, Six3/Xsix3, and the related factor Xsix6 (Xoptx2) (27, 30, 31). Our data suggest that the pathway used for ectopic eye formation after Xfz3 overexpression is similar to that used during endogenous eye development. The frequency of complete ectopic eye formation (11%) is similar to that obtained with overexpression of Xpax6 (27). In addition, the formation of ectopic pigment and effects on endogenous eye development were also observed with Xpax6 overexpression, arguing that Xfz3 acts in a common pathway with this gene to regulate eye development. Further supporting this conclusion, overexpression of Xfz3 was sufficient to promote ectopic expression of Xpax6, Xrx, and Xotx2. This ectopic expression occurred in regions of the embryo where the ectopic eyes formed, arguing that expression of these genes preceded and contributed to ectopic eye formation. In addition, disruption of signaling through Xfz3 by Nxfz3 blocked endogenous expression of Xpax6, Xrx, and Xotx2 in the anterior neural plate and blocked endogenous eye development. These data suggest that Xfz3 functions to control the expression of these eye regulatory genes. Our findings provide a clue to what is potentially required upstream of these transcription factors to initiate eye development. Whether the regulation by Xfz3 is direct or is acting in parallel overlapping pathways has yet to be determined.
The ectopic eyes were composed of cells that were overexpressing Xfz3, suggesting that it is a cell autonomous effect. Of interest was our observation that the ectopic eyes induced by Xfz3 overexpression appeared at or near the midline in proximity to the midbrain/hindbrain junction and often in the roof of the fourth ventricle. It is possible that there is a local source of wnts available to activate the Xfz3 receptor when Xfz3 is overexpressed. For example, Xwnt-1 and Xwnt-3A are highly expressed in the midbrain/hindbrain region of the developing Xenopus nervous system (32), and Xwnt-10 is expressed in the dorsal hindbrain (33). Alternatively, there may be requisite cooperating factors that are spatially restricted. For example, in some cases wnts can synergize with members of the fibroblast growth factor family to regulate patterning events (34). Finally, there may be a restricted subset of tissues competent to form eye tissue. Overexpression of Six3 or Six6 results in transformation of midbrain and rostral hindbrain tissue into retina, and it was suggested that this region of the nervous system is uniquely competent to form retinal tissue, perhaps because of endogenous expression of Xotx2 (30, 31). These possibilities have yet to be examined.
The observation that Xfz3 can initiate ectopic eye formation identifies wnt signaling as the first identified extracellular signaling pathway that regulates eye formation. Several wnts, including Xwnt-1, Xwnt-3A, and Xwnt-8, are expressed in the anterior neural plate in a region that overlaps the eye fields (32, 35). In addition, Xwnt-1 is much more potent than any other wnt ligand in synergizing with Xfz3 to promote both axis duplication and neural crest induction (M.A.D., J.-P. Saint-Jeannet, and P.S.K., unpublished observations). However, given the relative promiscuity of binding and the number of wnts present in the developing nervous system, it remains to be determined which wnt regulates eye development in vivo.
Frizzled activation can lead to signaling either through a canonical pathway involving β-catenin or noncanonical pathways that regulate planar cell polarity in Drosophila and possibly vertebrates (14–16), as well as calcium mobilization and protein kinase C activation in Xenopus and zebrafish (17). The signaling pathway used by Xfz3 to promote eye development has not yet been defined, although limited evidence points to the noncanonical planar cell polarity pathway. Overexpression of Xfz3 alone (unlike that of Xfz8) does not lead to axis duplication, a phenotype linked to activation of the canonical signaling pathway, although coexpression of Xfz3 with Xwnt1 can promote efficient axis duplication (M.A.D., J.-P. Saint-Jeannet, and P.S.K., unpublished observations). Expression of the closely related homolog Mfz3 in Xenopus embryos results in protein kinase C activation but not expression of siamois and Xnr3, which are downstream effectors in the canonical pathway (36). In addition, activation of the canonical wnt signaling pathway represses anterior neural development, arguing against mediation of the regulation of eye development by this pathway, although localized activation of the canonical pathway at later stages of development has not been examined.
We have used eight-cell RNA injection to overexpress a truncated form of Dsh (Dsh-ΔN), which preferentially activates the noncanonical planar cell polarity pathway (16). We found that 28% of the embryos (31/110) had dense ectopic pigment at or near the midline in the region of the hindbrain, reminiscent of the phenotype observed with Xfz3 overexpression. Conversely, injection of RNA encoding a truncated form of Dsh (Dsh-DEP+), which preferentially inhibits the noncanonical planar cell polarity pathway (16), resulted in reduced or missing eyes in 51% of injected embryos (52/101), similar to what was observed with Nxfz3 overexpression. These findings implicate the noncanonical planar cell polarity signaling pathway in the regulation of eye development, although this has yet to be confirmed.
In summary, our findings suggest a hitherto unsuspected role for wnt/frizzled signaling in regulating eye development and provide an example of a transmembrane receptor that is capable of promoting ectopic eye formation. Wnt-fz signaling may thus represent a critical link between extracellular patterning events and transcriptional regulation of early eye formation.
Acknowledgments
We thank Terry Van Raay for initiating the collaboration between our laboratories; Kathy Moore for advice on sectioning and immunohistochemistry; and Sheryl Scott, Villu Maricq, Joe Yost, and Kathy Moore for critical reading of the manuscript. We are grateful to Grant Mastick for the anti-Pax6 antibody. M.L.V. was supported by National Institutes of Health Grant EY12274 and by the Pew Scholars Program of the Pew Charitable Trusts. P.S.K. was supported by the Howard Hughes Medical Institute and a grant from the National Institutes of Health. M.A.D. was supported by the Life and Health Insurance Medical Research Fund.
Abbreviations
- Xfz3
Xenopus frizzled 3
- Nxfz3
an inhibitory form of Xfz3
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Eagleson G W, Harris W A. J Neurobiol. 1990;21:427–440. doi: 10.1002/neu.480210305. [DOI] [PubMed] [Google Scholar]
- 2.Hill R E, Favor J, Hogan B L, Ton C C, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 3.Mathers P H, Grinberg A, Mahon K A, Jamrich M. Nature (London) 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 4.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Development (Cambridge, UK) 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 5.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 6.Bhanot P, Brink M, Samos C H, Hsieh J C, Wang Y S, Macke J P, Andrew D, Nathans J, Nusse R. Nature (London) 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh J C, Rattner A, Smallwood P M, Nathans J. Proc Natl Acad Sci USA. 1999;96:3546–3551. doi: 10.1073/pnas.96.7.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang-Snyder J, Miller J R, Brown J D, Lai C J, Moon R T. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- 9.He X, Saint-Jeannet J P, Wang Y, Nathans J, Dawid I, Varmus H. Science. 1997;275:1652–1654. doi: 10.1126/science.275.5306.1652. [DOI] [PubMed] [Google Scholar]
- 10.Gradl D, Kuehl M, Wedlich D. Mech Dev. 1999;86:3–15. doi: 10.1016/s0925-4773(99)00129-x. [DOI] [PubMed] [Google Scholar]
- 11.Dorsky R I, Moon R T, Raible D W. Nature (London) 1998;396:370–373. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 12.Niehrs C. Trends Genet. 1999;15:314–319. doi: 10.1016/s0168-9525(99)01767-9. [DOI] [PubMed] [Google Scholar]
- 13.Shi D-L, Goisset C, Boucaut J-C. Mech Dev. 1998;70:35–47. doi: 10.1016/s0925-4773(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 14.Vinson C R, Conover S, Adler P N. Nature (London) 1989;338:263–264. doi: 10.1038/338263a0. [DOI] [PubMed] [Google Scholar]
- 15.Mlodzik M. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tada M, Smith J C. Development (Cambridge, UK) 2000;127:2227–2238. doi: 10.1242/dev.127.10.2227. [DOI] [PubMed] [Google Scholar]
- 17.Kühl M, Sheldahl L C, Park M, Miller J R, Moon R T. Trends Genet. 2000;16:279–283. doi: 10.1016/s0168-9525(00)02028-x. [DOI] [PubMed] [Google Scholar]
- 18.Deardorff M A, Klein P S. Mech Dev. 1999;87:229–233. doi: 10.1016/s0925-4773(99)00161-6. [DOI] [PubMed] [Google Scholar]
- 19.Nieuwkoop P D, Faber J. Normal Table of Xenopus laevis. New York: Garland; 1994. [Google Scholar]
- 20.Harland R M. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 21.Turner D L, Weintraub H. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 22.Adamus G, Zam Z S, Arendt A, Palczewski K, McDowell J H, Hargrave P A. Vision Res. 1991;31:17–31. doi: 10.1016/0042-6989(91)90069-h. [DOI] [PubMed] [Google Scholar]
- 23.Mastick G S, Davis N M, Andrews G L, Easter S S. Development (Cambridge, UK) 1997;124:1985–1997. doi: 10.1242/dev.124.10.1985. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch N, Harris W A. J Neurobiol. 1997;32:45–61. [PubMed] [Google Scholar]
- 25.Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, Boncinelli E. Development (Cambridge, UK) 1995;121:707–720. doi: 10.1242/dev.121.3.707. [DOI] [PubMed] [Google Scholar]
- 26.Moody S A, Kline M J. Anat Embryol. 1990;182:347–362. doi: 10.1007/BF02433495. [DOI] [PubMed] [Google Scholar]
- 27.Chow R L, Altmann C R, Lang R A, Hemmati-Brivanlou A. Development (Cambridge, UK) 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- 28.Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Development (Cambridge, UK) 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- 29.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 30.Loosli F, Winkler S, Wittbrodt J. Genes Dev. 1999;13:649–654. doi: 10.1101/gad.13.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernier G, Panitz F, Zhou X, Hollemann T, Gruss P, Pieler T. Mech Dev. 2000;93:59–69. doi: 10.1016/s0925-4773(00)00271-9. [DOI] [PubMed] [Google Scholar]
- 32.Wolda S L, Moody C J, Moon R T. Dev Biol. 1993;155:46–57. doi: 10.1006/dbio.1993.1005. [DOI] [PubMed] [Google Scholar]
- 33.Wolda S L, Moon R T. Oncogene. 1992;7:1941–1947. [PubMed] [Google Scholar]
- 34.McGrew L L, Hoppler S, Moon R T. Mech Dev. 1997;69:105–114. doi: 10.1016/s0925-4773(97)00160-3. [DOI] [PubMed] [Google Scholar]
- 35.Christian J L, Gavin B J, McMahon A P, Moon R T. Dev Biol. 1991;143:230–234. doi: 10.1016/0012-1606(91)90073-c. [DOI] [PubMed] [Google Scholar]
- 36.Sheldahl L C, Park M, Malbon C C, Moon R T. Curr Biol. 1999;9:695–698. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]