Abstract
Homeodomain only protein x (Hopx) is an unusual homeodomain protein that has diverse effects on cardiac growth. Manipulation of Hopx function in murine models is associated with cardiac hypertrophy, dilation and fibrosis. In the present study, we examined the expression profile of Hopx in various models of pathologic cardiac hypertrophy and failure. Hopx expression is significantly reduced in neonatal rat cardiac myocytes after α/β adrenergic receptor agonist treatment. Cardiac hypertrophy and failure induced by transaortic constriction in mice causes marked down-regulation of Hopx expression. Interestingly, HOPX expression was significantly reduced in hearts of humans with end stage heart failure when compared to non-failing control hearts, and HOPX levels remains low after LVAD support. Our findings suggest that HOPX/Hopx expression is reduced in multiple examples of human and murine cardiac hypertrophy and failure.
Keywords: Hopx, hypertrophy, myocyte, heart failure, LVAD
1. Introduction
In response to the pathological conditions, including hemodynamic and neurohormonal stress, the heart undergoes maladaptive growth to sustain function. However, hypertrophic growth can lead to energy supply-demand mismatch, loss of cardiomyocytes, progressive cardiac dysfunction, and heart failure, which is a leading cause of morbidity and mortality in the United States [1]. Current therapies primarily target hemodynamic responses rather than intrinsic cellular defects of the ailing myocyte. Several transcription factors, co-factors, and associated signaling pathways have been implicated in the pathogenesis of cardiac hypertrophy and failure in murine models [2]. Due to limitations of assessment in human subjects, less is known about roles of these transcription factors and signaling pathways in human heart failure.
The homeodomain only protein x (Hopx) is a small 73 amino acid protein that is largely composed of a 60 amino acid motif homologous to the homeodomain of Hox transcription factors[3]. Unlike Hox homeodomains, Hopx is unable to bind DNA as it lacks certain conserved amino acid residues that are required for protein-DNA interactions[3, 4]. However, loss- and gain-of function murine models have shown that Hopx functions as a transcription co-factor to modulate cardiac specific gene program and thereby cardiac growth. Loss of Hopx function results in cardiac hypertrophy and activation of “fetal gene” program in adult mice[4]. Hopx regulates cardiac growth, in part, by directly interacting with serum response factor (SRF) and repressing its transcriptional activity [3, 4]. Interestingly, gain of Hopx function in transgenic mice is sufficient to induce cardiac hypertrophy by recruiting histone deacetylase (Hdac) 2 activity to inhibit anti-hypertrophic gene transcription[5]. Though previous studies have described Hopx as a regulator of cardiac hypertrophy, its expression pattern in murine and human cardiac hypertrophy and failure is not known. Here we demonstrate that Hopx/HOPX expression is significantly down-regulated in response to hypertrophic stimuli, both in vitro and in vivo, and in end-stage heart failure patients.
2. Materials and methods
2.1. Primary cardiomyocyte cultures
Neonatal rat ventricular cardiac myocytes (NRCM) were isolated and cultured as described previously [6]. Cells were treated with 1 μM angiotensin II (Sigma) or 10nm–100 μM isoproterenol (Sigma).
2.2. Animal models
All mice were maintained in the animal facilities at the University of Pennsylvania and protocols were approved by the institutional animal care and use committee (IACUC). Transverse aortic constriction (TAC) was performed using 10–12 weeks old CD1 mice (Charles River) as described elsewhere [7].
2.3. Quantitative real time PCR
Quantitative real time PCR (qRT-PCR) was performed as described previously [8]. Briefly, total RNA was isolated from cultured NRCM treated with or without isoproterenol or angiotensin II, or from dissected mouse heart ventricles using Trizol (Invitrogen). RNA was reverse-transcribed using random hexamers and the Superscript First Strand Synthesis Kit (Invitrogen). Gene expression was then evaluated by qRT-PCR (ABI PRISM 7900) using the SYBR Green (Applied Biosystems). Signals were normalized to their corresponding GAPDH controls and the ratios expressed as fold changes compared to wild-type controls. PCR conditions and primer set sequences are available upon request.
2.4. Human myocardial samples
Human myocardium was obtained from patients undergoing cardiac transplantation (failing and left ventricular assist device (LVAD)-supported) or from donor hearts deemed unsuitable for transplantation (non-failing) as described elsewhere [9]. Institutional Review Board approval was obtained for these studies, and prospective or retrospective informed consent for research use of donated heart tissue was obtained from all living patients.
2.5. Microarray analysis
The sample preparation, processing procedure and analysis was performed according to the Affymetrix GeneChip Expression Analysis manual as described elsewhere [9].
2.6. Statistics
The statistical analysis between different groups was performed by Student's two-tailed t-test. Differences were considered significant at a P-value < 0.05.
3. Results
3.1. Hypertrophy decreases Hopx expression in cultured myocytes
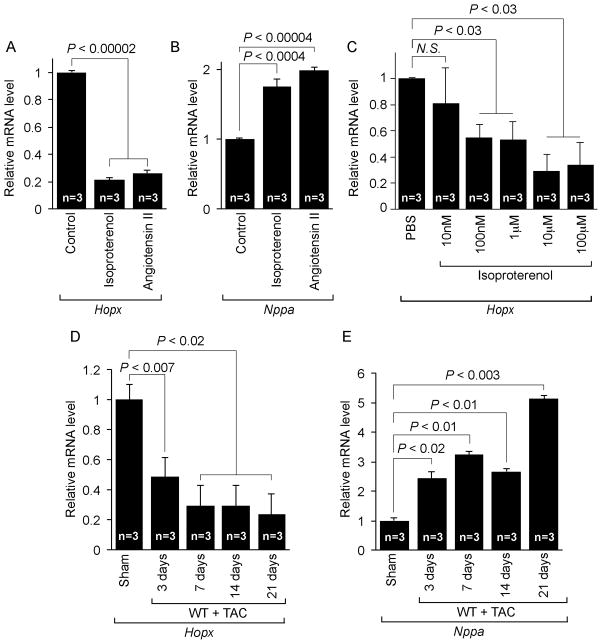
We determined the expression levels of Hopx in cultured NRCMs in response to the hypertrophic agonists isoproterenol and angiotensin II. Hopx mRNA was down-regulated 70–80% by each of these agonists (Figure 1A). Nppa expression, a classical marker of cardiac hypertrophy, was up-regulated by ~2 fold in response to either isoproterenol or angiotensin II (Figure 1B). Interestingly, Hopx mRNA was down-regulated in a dose-dependent manner in response to isoproterenol (Figure 1C). In a murine model of cardiac hypertrophy induced by transverse aortic constriction (TAC), Hopx mRNA was ~50% reduced within three days of TAC induced hypertrophy (Figure 1D) when compared to sham operated control animals. Notably, Hopx expression was further reduced by 7 days of aortic constriction, and remained low up to 21 days (Figure 1D) when fibrosis and heart failure is evident (not shown). Nppa expression was up-regulated in response to TAC induced hypertrophy (Figure 1E), as expected. Together, these data suggest that hypertrophic stimuli reduce Hopx expression in vivo and in vitro.
Figure 1.
Downregulation of Hopx expression in response to hypertrophic stimuli in vitro and in vivo: (A-C) Hopx or Nppa transcripts were detected by qRT-PCR from cultured neonatal rat cardiomyocytes treated with or without isoproterenol or angiotensin II for 48 hours (mean ± SEM, n=3). Gapdh serves as control. (D-E) Transcripts for Hopx and Nppa were detected in adult mice hearts at day 0, 3, 7, 14, and 21 after sham (n=3) or TAC (n=3) surgery by qRT-PCR.
3.2. HOPX expression is down-regulated in human heart failure
To understand the HOPX expression pattern in human heart failure, we quantified expression using a large bank of human heart tissue. We obtained the left ventricular myocardium from patients with end-stage heart failure, non-failing organ donors with normal ventricular function, and LVAD-supported patients with heart failure at the time of heart transplantation [9]. Microarray analysis of these samples showed dysregulation of approximately 200 genes with a 2-fold or greater difference between non-failing and failing heart samples [9]. We observed approximately 4-fold down-regulation in HOPX expression in failing hearts compared to non-failing hearts (Figure 2A). This was associated with increased expression of Nppa (data not shown). Despite regression of pathologic hypertrophy in LVAD-supported hearts, HOPX expression remained substantially lower than in non-failing controls (Figure 2A). Differential HOPX expression in failing and LVAD supported hearts was verified by quantitative real-time PCR (Figure 2B).
Figure 2.
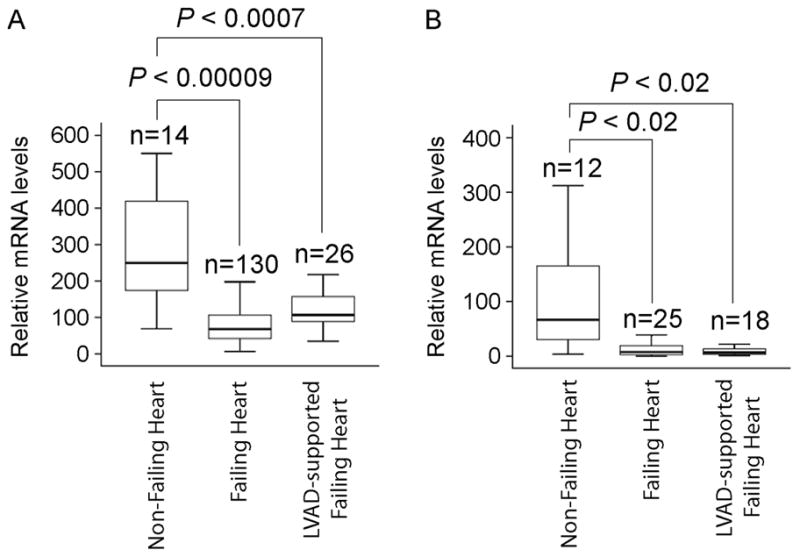
HOPX mRNA is downregulated in end-stage heart failure patients: Box plot represents HOPX transcript levels, detected by either microarray analysis (A) or qRT-PCR (B), in non-failing, failing and LVAD supported human hearts.
4. Discussion
Several transcription factors and co-factors, including SRF, Nkx2-5, Gata4, Nfat, Hdacs and Hopx, regulate cardiac hypertrophic gene programs in murine models [1]. Recently, MEF2, NKX2-5, and GATA4 have been associated with common forms of human heart failure [10]. Though we have previously described the role of Hopx in cardiac hypertrophy and early heart failure in mice, here we determined that HOPX expression is reduced in advanced human heart failure, consistent with a previous report [11]. We also show that murine Hopx expression is down-regulated in response to hypertrophic stimuli in vivo and in vitro.
Increased SRF activity is important for the activation of pro-hypertrophic gene programs and induction of the cardiac hypertrophy [12]. Hopx negatively regulates SRF dependent transcriptional activity by interacting directly with SRF and by promoting its dissociation from DNA [3, 4]. Our findings suggest that down-regulation of Hopx/HOPX expression, in response to hypertrophic stimuli, could be an important event facilitating activation of SRF dependent pro-hypertrophic gene expression. Similarly, GATA4 has been associated with human cardiac hypertrophy and end-stage heart failure [10]. Increased Gata4 dependent transcriptional activity can activate a pro-hypertrophic gene program [13]. Recently, Hopx has been shown to inhibit Gata4 dependent transcription by recruiting Hdac2 [14]. Loss of Hopx function increases Gata4 activity in mice, suggesting that decreased HOPX expression in human heart failure patients might augment Gata4 dependent pro-hypertrophic gene program. Additional studies will be necessary to determine if augmentation of Hopx activity could affect the progression of cardiac hypertrophy and failure in animal models and human patients.
Acknowledgments
This work was supported by the American Heart Association DeHaan Myogenesis Center, the WW Smith Charitable Trust, and the NIH (RO1 HL071546 to JAE, R01 HL088577 to TPC, R01 AG17022 to KBM, and K99 HL098366 to CMT)
Footnotes
Disclosure: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006 Aug;7(8):589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 2.Putt ME, Hannenhalli S, Lu Y, Haines P, Chandrupatla HR, Morrisey EE, et al. Evidence for coregulation of myocardial gene expression by MEF2 and NFAT in human heart failure. Circ Cardiovasc Genet. 2009 Jun;2(3):212–9. doi: 10.1161/CIRCGENETICS.108.816686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F, Kook H, Milewski R, Gitler AD, Lu MM, Li J, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002 Sep 20;110(6):713–23. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 4.Shin CH, Liu ZP, Passier R, Zhang CL, Wang DZ, Harris TM, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002 Sep 20;110(6):725–35. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 5.Kook H, Epstein JA. Hopping to the beat. Hop regulation of cardiac gene expression. Trends Cardiovasc Med. 2003 Oct;13(7):261–4. doi: 10.1016/s1050-1738(03)00107-5. [DOI] [PubMed] [Google Scholar]
- 6.Kubo H, Jaleel N, Kumarapeli A, Berretta RM, Bratinov G, Shan X, et al. Increased cardiac myocyte progenitors in failing human hearts. Circulation. 2008 Aug 5;118(6):649–57. doi: 10.1161/CIRCULATIONAHA.107.761031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007 Mar;13(3):324–31. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 8.Trivedi CM, Lu MM, Wang Q, Epstein JA. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem. 2008 Sep 26;283(39):26484–9. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA, Bednarik D. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ Res. 2005 Mar 18;96(5):592–9. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 10.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, et al. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006 Sep 19;114(12):1269–76. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 11.Torrado M, Lopez E, Centeno A, Medrano C, Castro-Beiras A, Mikhailov AT. Myocardin mRNA is augmented in the failing myocardium: expression profiling in the porcine model and human dilated cardiomyopathy. J Mol Med. 2003 Sep;81(9):566–77. doi: 10.1007/s00109-003-0470-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Azhar G, Chai J, Sheridan P, Nagano K, Brown T, et al. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am J Physiol Heart Circ Physiol. 2001 Apr;280(4):H1782–92. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- 13.Molkentin JD, Olson EN. GATA4: a novel transcriptional regulator of cardiac hypertrophy? Circulation. 1997 Dec 2;96(11):3833–5. [PubMed] [Google Scholar]
- 14.Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, et al. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell. 2010 Sep 14;19(3):450–9. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]