Abstract
Electron-transfer dissociation (ETD) with supplemental activation of the doubly charged deamidated tryptic digested peptide ions allows differentiation of isoaspartic acid and aspartic acid residues using c + 57 or z• − 57 peaks. The diagnostic peak clearly localizes and characterizes the isoaspartic acid residue. Supplemental activation in ETD of the doubly charged peptide ions involves resonant excitation of the charge reduced precursor radical cations and leads to further dissociation, including extra backbone cleavages and secondary fragmentation. Supplemental activation is essential to obtain a high quality ETD spectrum (especially for doubly charged peptide ions) with sequence information. Unfortunately, the low-resolution of the ion trap mass spectrometer makes detection of the diagnostic peak for the aspartic acid residue difficult due to interference with side-chain loss from arginine and glutamic acid residues.
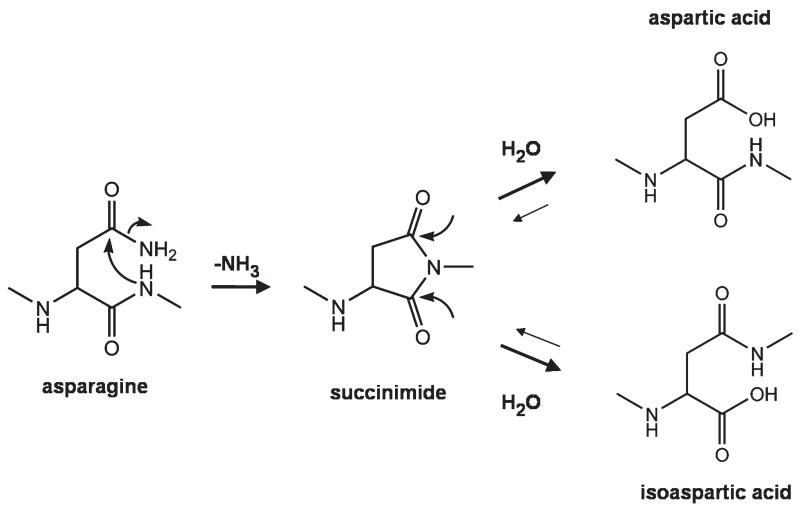
Deamidation of asparagine (N) and isomerization of aspartic acid (D) to form a mixture of aspartic and isoaspartic acid (isoD) are nonenzymatic post-translational modifications that occur spontaneously in proteins [1]. Deamidation occurs through a cyclic succinimide intermediate formed by nucleophilic attack of the side-chain carbonyl by the backbone amide (Scheme 1) [2, 3]. This intermediate is not stable in aqueous solution and undergoes hydrolysis, resulting in a mixture of aspartic and isoaspartic acid typically in a 1:3 ratio in favor of the isoaspartic acid form [4]. Deamidation half-life ranges from 1 to 500 d and the rate of deamidation is significantly affected by the steric hindrance of the C-terminal amino acid of aspartic acid [5, 6] and the higher order structure of the protein. It was found that deamidation of peptides containing-NG- would be the fastest among the 20 common amino acids as glycine (G) has the smallest steric hindrance. Isomerization of aspartic acid has been found, in some cases, to have significant impact on biological activity as the tertiary structure is disrupted by one methylene group (CH2) shifted from the side chain to the protein backbone. These two structural changes dramatically alter the folding of the protein and are postulated to be correlated with initiation of Alzheimer’s disease [7-9].
Scheme 1.
The standard mechanism for deamidation of asparagine and isomerization of aspartic acid residues.
Analytical techniques that can be used in determination of deamidation of aspartic acid include gel electrophoresis [10], Edman degradation [7], high-performance liquid chromatography (HPLC) [7], and mass spectrometry (MS) [11]. Gel electrophoresis can be used to detect a deamidated protein as the isoelectric point (pI) changes, however, it generally cannot be used to differentiate aspartic and isoaspartic acid as these two isomers have similar reactivity. Edman degradation can distinguish the isomers but the protein mixture has to be separated by HPLC before analysis. However, HPLC has the drawback that it requires a relatively high concentration of sample to obtain a signal.
In addition, standard peptides containing aspartic and isoaspartic acid are needed to perform the control experiments in HPLC. Among the above techniques, mass spectrometry is the most sensitive and has the shortest analysis time for distinguishing aspartic acid from isoaspartic acid. With the use of conventional dissociation techniques in MS such as collisionally activated dissociation (CAD), it is difficult to differentiate the isomers [12]. Electron capture dissociation (ECD) used in Fourier transform ion cyclotron resonance mass spectrometers (FTICRMS) has demonstrated that aspartic and isoaspartic acid can be differentiated by generation of diagnostic ion(s) unique to each form [13-16].
In ECD, the captured electron initiates N–Cα backbone cleavages forming c and z• ions normally. Since isoaspartic acid does not contain any N–Cα bonds, its fragmentation under ECD conditions is different from that of aspartic acid. A diagnostic ion pair, cl-n + 57 (or cl-n• + 58) and zn• − 57 (l is the length of the peptide, n is isoaspartic acid), is observed as backbone fragment(s) for isoaspartic acid and, hence, its identity and position(s) can be determined [12]. Due to the similarity between electron-transfer dissociation (ETD) and ECD, differentiation of aspartic and isoaspartic acid using ETD is also possible by detecting the presence of c + 57 and z• − 57 peaks [17]. However, recently, Hayley et al. have shown that four out of eight peptides in their study did not give useful ETD spectra in positive ion mode [18]. For the rest of the peptides, c + 57 and z• − 57 ions were detectable but in very low abundance, and therefore there are difficulties to characterize aspartic and isoaspartic acid in doubly charged peptides. Difficulties with ETD of doubly charged precursor ions have been reported, [19] and it was demonstrated that the use of supplemental activation in ETD can solve this problem [20, 21]. Therefore the use of supplementary activation in ETD may solve the problem of detection of c + 57 and z• − 57 ions in deamidated peptides.
The purpose of this communication is to show that with the use of supplemental activation in ETD (especially for doubly charged precursor ions), differentiation of aspartic and isoaspartic acid in tryptic digested peptides is facilitated.
Methods
All chemicals and proteins were purchased from Sigma-Aldrich (St. Louis, MO, USA). Bovine serum albumin (BSA), calmodulin, and lysozyme were digested with trypsin at 1:50 (wt/wt) in 100 μM ammonium bicarbonate (pH 8.5) for 12 h at 37 °C. Five percent formic acid was added to quench the reaction. The solution mixtures were freeze-dried to remove ammonia and then dissolved to a final concentration of 2.5 μM in 49.5:49.5:1 (vol:vol:vol) methanol:water:formic acid for ESI-ETD experiments.
All ETD experiments were performed on a 3D high capacity ion trap mass spectrometer (HCT Ultra; Bruker Daltonics). Peptides were directly infused into the source at a flow rate of 4 μL/min and ionized at 4000 V with skimmer voltage at 40 V. For ETD experiments, fluoranthene radical anions generated in the negative chemical ionization (nCI) source were tuned to maximize output of ETD reagent ion (202 m/z), with reagent ion charge control (ICC) 5e + 05, ionization energy 70 eV, emission current 4.5 μA, and a low mass cut-off at 210 m/z to remove remaining reactant. In each cycle, the peptide ions were reacted with ~5e + 05 fluoranthene ions for 400 ms with or without supplemental activation (smart decomposition: z = 2, z = 3, or off).
Results and Discussion
Figure 1 shows the ETD spectra (doubly charged precursor ions) of the deamidated tryptic digested peptide 92VFDKDGDGYISAAELR107 (D in bold font indicates the aspartic or isoaspartic residue obtained from deamidation and hydrolysis of asparagine residue) from calmodulin with supplemental activation (Figure 1a), and without supplemental activation (Figure 1b). In ETD spectra of [M + 2H]2+, backbone fragments are mainly z• ions instead of complementary c and z• ions. This is because arginine or lysine (basic amino acid) is located at the C-terminus of the tryptic digested peptide, thus, one charge is retained at the C-terminal basic amino acid the other being reduced by the electrons. Therefore, the abundance of z• ions are generally much higher than that of c ions in spite of decreased stability. A wide range of z• ions ( to ) but only four c ions were observed in Figure 1a while only three z• ions ( to ) and no c ions were observed in Figure 1b. Thus, the sequence coverage and abundance of fragment peaks were dramatically increased with the use of supplemental activation. In addition, the diagnostic z10• − 57 ion yielding information on the isoaspartic acid residue position was only detected in Figure 1a. The M-60 Da product ion, which is diagnostic of the presence of the aspartic acid residues, showed interference from the M-59 Da even-electron arginine/odd-electron glutamic acid side-chain cleavage fragments. The corresponding to cleavage N-terminal to the aspartic or isoaspartic acid residue appears to have similar abundance when compared to and corresponding to cleavage N-terminal to the aspartic acid residues.
Figure 1.

Electron-transfer dissociation (ETD) spectra of the doubly charged deamidated tryptic digested peptide 92VFDKDGDGYISAAELR107 from calmodulin (a) with supplemental activation and (b) without supplemental activation. Bold denotes isoaspartic acid diagnostic ions.
The ETD spectra (doubly charged precursor ions) of the deamidated tryptic digested peptide 421LGEYGFQDALIVR433 from BSA and 46NTDGSTDYGILQIDSR61 from lysozyme with and without supplemental activation are included in the supplementary information, which can be found in the electronic version of this article. These experimental results are similar to that of ETD of doubly charged deamidated tryptic digested peptide from calmodulin as discussed above. With the use of supplemental activation, backbone fragment (mainly from z• ions) abundance and sequence coverage significantly increased, as well as generation of the diagnostic cl-n + 57 or zn• − 57 ion. A possible mechanism of the formation of cl-n + 57 or zn• − 57 ion was proposed by Cournoyer et al. [13] In addition, the intensities of z• ions corresponding to cleavage N-terminal to the isoaspartic acid and aspartic acid residues were relatively high compared with other backbone positions.
Figure 2 shows the ETD spectra (triply charged precursor ions) of the deamidated tryptic digested peptide 127EADIDGDGQVDYEEFVQMMTAK148 from calmodulin with supplemental activation (Figure 2a), and without supplemental activation (Figure 2b). In ETD of [M + 3H]3+, besides the typical complementary c and z• ion pairs, minor backbone fragments such as a, b, x, and y ions were also detected. The use of supplemental activation has not only produced the diagnostic c10 + 57 ion, but also increased the intensities of all fragment peaks by resonant excitation of the reduced molecular ion [M + 2H]2+• or [M + 3H]+•• (base peak shifted from [M + 2H]2+ in Figure 2b to [M + H]+ in Figure 2a). Comparison of Figure 1b and Figure 2b shows that ETD of [M + 3H]3+ generated more different types of backbone fragments (with low abundance) and the sequence coverage was relatively high, suggesting that the internal energy of the charge-reduced molecular ion [M + 2H]2+• or [M + 3H]+•• was high enough to induce additional backbone cleavages. Presumably, this observed increase in fragment ion number and abundance is related to the increased Coulombic repulsion inherent in more highly charged species, which results in a more unfolded structure and in fragments that can experience some Coulombic energy release. However, due to the limited mass accuracy of the ion trap instrument used for these ETD studies, definitive assignment of several odd- or even-electron side-chain losses from the z• ions or [M + 2H]+• was difficult due to overlap with some of the singly and doubly charged backbone fragments.
Figure 2.

Electron-transfer dissociation (ETD) spectra of the triply charged deamidated tryptic digested peptide 128EADIDGDGQVDYEEFVQMMTAK149 from calmodulin (a) with supplemental activation and (b) without supplemental activation. Bold denotes isoaspartic acid diagnostic ions.
These experiments show that the effect of supplemental activation in ETD of the doubly charged precursor ions was more profound than that of the triply charged precursor ions. With the use of supplemental activation in ETD, more backbone fragments were observed. It is tentatively believed that the neutralization energy released by the reaction between the transferred electron and charge is alone often insufficient to induce further dissociation or separate the hydrogen bonded fragment ion complex. When this non-dissociative electron-transfer product ([M + 2H]+•) was resonantly excited, it yielded backbone fragments or side-chain loss from [M + 2H]+• (shown in Figure 1a).
Conclusions
Electron-transfer dissociation with supplemental activation demonstrated that for doubly charged peptide ions, supplemental activation is essential to obtain the diagnostic c + 57 or z• − 57 peaks needed to differentiate aspartic and isoaspartic acids in deamidated, digested peptide ions. These diagnostic peaks clearly define both the presence and position of isoaspartic acid residues. With the use of supplemental activation, the reduced precursor ion ([M + 2H]+•) was resonantly excited and underwent further dissociation. As a result, the abundance of backbone fragments dramatically increased and the sequence coverage was significantly improved from typically ~50% to ~85%. For ETD of triply charged peptide ions, supplemental activation was also necessary to generate the c + 57 or z• − 57 ions as well as increasing the backbone fragment intensities. The low-resolution of the ion trap instrument makes detection of M-60 Da (the aspartic acid residue’s diagnostic peak) difficult due to interference with side-chain fragment ions (59 Da) from arginine and glutamic acid residues. In addition, it was difficult to distinguish the charge states of some of the backbone fragments in ETD of high charge state precursor ions (e.g., [M + 3H]3+) due to the poor resolution of the ion trap mass spectrometer.
Supplementary Material
Acknowledgments
The authors acknowledge that this research was sponsored in part by the Warwick Centre for Analytical Science, The National Institutes of Health (GM078293), The Warwick Department of Chemistry, and the Research Grants Council of the HKSAR, China (reference no. 400207). W.Y.K.C. is grateful for the Chinese University of Hong Kong studentship.
Footnotes
Appendix A
Supplementary Material
Supplementary material associated with this article may be found in the online version at doi:10.1016/j.jasms.2010.02.002.
References
- 1.Robinson NE, Robinson AB. Molecular Clocks: Deamidation of Asparaginyl and Glutaminyl Residues in Peptides and Proteins. Althouse Press; Cave Junction, OR: 2004. [Google Scholar]
- 2.Clarke S. Propensity for Spontaneous Succinimide Formation from Aspartyl and Asparaginyl Residues in Cellular Proteins. Int. J. Pept. Protein Res. 1987;30:808–821. doi: 10.1111/j.1399-3011.1987.tb03390.x. [DOI] [PubMed] [Google Scholar]
- 3.Radkiewics JL, Zipse H, Clarke S, Houk KN. Accelerated Racemization of Aspartic Acid and Asparagine Residues Via Succinimide Intermediates: An Ab Initio Theoretical Exploration of Mechanism. J. Am. Chem. Soc. 1996;118:9148–9155. [Google Scholar]
- 4.Geiger T, Clarke S. Deamidation, Isomerization, and Racemization at Asparaginyl and Aspartyl Residues in Peptides. J. Biol. Chem. 1987;262:785–795. [PubMed] [Google Scholar]
- 5.Robinson NE, Robinson AB. Molecular Clocks. Proc. Natl. Acad. Sci. U.S.A. 2001;98:944–949. doi: 10.1073/pnas.98.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson NE, Robinson AB. Prediction of Protein Deamidation Rates from Primary and Three-Dimensional Structure. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roher AE, Lowenson JD, Clarke S, Wolkow C, Wang R, Cotter RJ, Reardon IM, Zurcher-Neely HA, Heinrikson RL, Ball MJ. Structural Alterations in the Peptide Backbone of β-Amyloid Core Protein may account for its Deposition and Stability in Alzheimer’s Disease. J. Biol. Chem. 1993;268:3072–3083. [PubMed] [Google Scholar]
- 8.Shimizu T, Watanabe A, Ogawara M, Mori H, Shirasawa T. Isoaspartate Formation and Neurodegeneration in Alzheimer’s Disease. Arch. Biochem. Biophys. 2000;381:225–234. doi: 10.1006/abbi.2000.1955. [DOI] [PubMed] [Google Scholar]
- 9.Sargaeva NP, Lin C, O’Connor PB. Identification of Aspartic and Isoaspartic Acid Residues in Amyloid β Peptides, Including Aβ1–42, Using Electron-Ion Reactions. Anal. Chem. 2009;81:9778–9786. doi: 10.1021/ac901677t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu YR, Chang WC, Mendiza EA, Hara S, Chow DT, Mann MB, Langley KE, Lu HS. Selective Deamidation of Recombinant Human Stem Cell Factor During In Vitro Aging; Isolation, and Characterization of the Aspartyl and Isoaspartyl Homodimers and Heterodimers. Biochemistry. 1998;37:2251–2262. doi: 10.1021/bi972372z. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann W, Schlosser A, Erben G, Pipkorn R, Bossemeyer D, Kinzel V. Analysis of Isoaspartate in Peptides by Electrospray Tandem Mass Spectrometry. Protein Sci. 2000;9:2260–2268. doi: 10.1110/ps.9.11.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schindler P, Muller D, Marki W, Grossenbacher H, Richter WJ. Characterization of a β-Asp33 Isoform of Recombinant Hirudin Sequence Variant 1 by Low-Energy Collision-Induced Dissociation. J. Mass Spectrom. 1996;31:967–974. doi: 10.1002/(SICI)1096-9888(199609)31:9<967::AID-JMS381>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 13.Cournoyer JJ, Pittman JL, Ivlevaver AB, Fallows E, Waskell L, Costello CE, O’Connor PB. Deamidation: Differentiation of Aspartyl from Isoaspartyl Products in Peptides by Electron Capture Dissociation. Protein. Sci. 2005;14:452–463. doi: 10.1110/ps.041062905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cournoyer JJ, Lin C, O’Connor PB. Detecting Deamidation Products in Proteins by Electron Capture Dissociation. Anal. Chem. 2006;78:1264–1271. doi: 10.1021/ac051691q. [DOI] [PubMed] [Google Scholar]
- 15.Cournoyer JJ, Lin C, Bowman MJ, O’Connor PB. Quantitating the Relative Abundance of Isoaspartyl Residues in Deamidated Proteins by Electron Capture Dissociation. J. Am. Soc. Mass Spectrom. 2007;18:48–56. doi: 10.1016/j.jasms.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Li XJ, Cournoyer JJ, Lin C, O’Connor PB. Use of 18O Labels to Monitor Deamidation during Protein and Peptide Sample Processing. J. Am. Soc. Mass Spectrom. 2008;19:855–864. doi: 10.1016/j.jasms.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connor PB, Cournoyer JJ, Pitteri SJ, Chrisman PA, McLuckey SA. Differentiation of Aspartic and Isoaspartic Acids Using Electron Transfer Dissociation. J. Am. Soc. Mass Spectrom. 2006;17:15–19. doi: 10.1016/j.jasms.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Andreazza HJ, Wang TF, Bagley CJ, Hoffmann P, Bowie JH. Negative Ion Fragmentations of Deprotonated Peptides. The Unusual Case of isoAsp: A Joint Experimental and Theoretical Study. Comparison with Positive Ion Cleavages. Rapid Commun. Mass Spectrom. 2009;23:1993–2002. doi: 10.1002/rcm.4107. [DOI] [PubMed] [Google Scholar]
- 19.Pitteri SJ, Chrisman PA, Hogan JM, McLuckey SA. Electron Transfer Ion/Ion Reactions in a Three-Dimensional Quadrupole Ion Trap: Reactions of Doubly and Triply Protonated Peptides with SO2. Anal. Chem. 2005;77:1831–1839. doi: 10.1021/ac0483872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Supplemental Activation Method for High-Efficiency Electron-Transfer Dissociation of Doubly Protonated Peptide Precursors. Anal. Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domon B, Bodenmiller B, Carapito C, Hao ZQ, Huehmer A, Aebersold R. Electron Transfer Dissociation in Conjunction with Collision Activation to Investigate the Drosophila melanogaster Phosphoproteome. J. Proteome Res. 2008;8:2633–2639. doi: 10.1021/pr800834e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.