Abstract
CBS domains are defined as sequence motifs that occur in several different proteins in all kingdoms of life. Although thought to be regulatory, their exact functions have been unknown. However, their importance was underlined by findings that mutations in conserved residues within them cause a variety of human hereditary diseases, including (with the gene mutated in parentheses): Wolff-Parkinson-White syndrome (γ2 subunit of AMP-activated protein kinase); retinitis pigmentosa (IMP dehydrogenase-1); congenital myotonia, idiopathic generalized epilepsy, hypercalciuric nephrolithiasis, and classic Bartter syndrome (CLC chloride channel family members); and homocystinuria (cystathionine β-synthase). AMP-activated protein kinase is a sensor of cellular energy status that is activated by AMP and inhibited by ATP, but the location of the regulatory nucleotide-binding sites (which are prime targets for drugs to treat obesity and diabetes) was not characterized. We now show that tandem pairs of CBS domains from AMP-activated protein kinase, IMP dehydrogenase-2, the chloride channel CLC2, and cystathionine β-synthase bind AMP, ATP, or S-adenosyl methionine,while mutations that cause hereditary diseases impair this binding. This shows that tandem pairs of CBS domains act, in most cases, as sensors of cellular energy status and, as such, represent a newly identified class of binding domain for adenosine derivatives.
Introduction
CBS domains were originally identified as sequence motifs of approximately 60 amino acids that occur in CBS and several other proteins, in all organisms from archaea to humans (1). Although their functions were unknown, their importance was emphasized by findings that point mutations within them cause several hereditary diseases in humans. These include (with the protein/gene affected in parentheses): homocystinuria (cystathionine β-synthase [ref. 2]); retinitis pigmentosa (IMP dehydrogenase-1 [refs. 3, 4]); congenital myotonia, idiopathic generalized epilepsy, hypercalciuric nephrolithiasis, and classic Bartter syndrome (chloride channels CLC1, CLC2, CLC5, and CLCKB, respectively [refs. 5–8]); and Wolff-Parkinson-White syndrome (WPWS) (γ2 subunit of AMP-activated protein kinase [refs. 9–12]). Although not yet reported in humans, mutations in the CBS domains of the AMP-activated protein kinase (AMPK) γ3 subunit cause an abnormally high glycogen content in skeletal muscle of pigs (13).
AMPK is a cellular energy sensor that is activated by AMP and inhibited by high concentrations of ATP (14). Once activated by cellular stresses that cause ATP depletion, the kinase switches on catabolic pathways that generate ATP and switches off ATP-consuming processes. As well as maintaining energy balance at the cellular level, it is now becoming clear that AMPK plays an important role in regulating whole-body energy storage and expenditure. The drug 5-aminoimidazole-4-carboxamide (AICA) riboside, which activates AMPK, alleviates metabolic defects of type 2 diabetes and the metabolic syndrome in animal models (15–18). The kinase is activated by exercise in muscle (19), stimulating glucose and fatty acid oxidation (20), and may be partly responsible for the protective effects of exercise in the development of obesity and type 2 diabetes. It is also activated by the antidiabetic drugs metformin and rosiglitazone (21–23), and by the adipocyte-derived hormones leptin and adiponectin (24, 25). AMPK activators are now recognized as prime candidates for new drugs to treat type 2 diabetes, obesity, and the metabolic syndrome (26, 27), but the locations of the regulatory AMP- and ATP-binding sites (the most likely binding sites for activators) have not been characterized.
AMPK exists as heterotrimeric complexes comprising catalytic α subunits and regulatory β and γ subunits (14). CBS domains invariably occur as tandem pairs, but the three γ subunit isoforms of AMPK, and their homologues in lower eukaryotes, are unique among eukaryotic proteins in having four tandem domains, although other examples are found in archaeal genomes. We have previously shown, using a photolabile AMP analogue, that the γ subunits are involved in binding of the nucleotide (28). Since the four CBS domains are the only regions conserved between the three γ isoforms, we hypothesized that they form the allosteric nucleotide-binding sites. In this paper we have examined the hypothesis that pairs of CBS domains form binding sites for adenosine-containing ligands, and that this is disrupted by disease mutations.
Methods
DNA cloning and expression.
PCDNA plasmids encoding rat γ1 (29) and human γ2 and γ3 (28) were used as templates for PCR. DNAs encoding all four CBS domains (CBS1-4) from γ1 (residues 42–324), γ2 (residues 274–569), and γ3 (residues 197–479), and the N-terminal domain pair from γ2 (CBS1-2, residues 274–410) or the C-terminal domain pair from γ2 (CBS3-4, residues 430–556), were amplified using the Pfu Turbo PCR kit (Stratagene, La Jolla, California, USA). The primers were designed to amplify DNA encoding the designated amino acids but also contained 5′-BamHI and 3′-XhoI ends. The PCR products were cloned into pGEX-KG using those restriction sites. For IMP dehydrogenase-2 (IMPDH2), DNAs encoding residues 112–232 or the full-length protein (residues 1–514) were amplified using human liver cDNA (BD Biosciences Clontech, Oxford, United Kingdom) as template, and cloned into pHAT20 (BD Biosciences Clontech) using 5′-AgeI and 3′-KpnI sites added to the primers. For the other constructs, DNAs encoding residues 582–840 of CLC2, or 416–551 of cystathionine β-synthase, were amplified using human liver cDNA as template, and cloned into pGEX-KG as described above. All mutations were created using the QuikChange Site-Directed Mutagenesis system (Stratagene).
Bacterial expression and purification of proteins.
Luria-Bertani medium (1 liter) containing ampicillin (50 μg/μl) was inoculated with overnight cultures (10 ml) of Escherichia coli BL21 (DE3) containing the appropriate construct. Cultures were grown at 37°C until the absorbance at 600 nm reached 0.4, after which isopropyl thiogalactoside (1 mM) was added to induce expression. Cells were recovered by centrifugation 3 hours later, and cell pellets were resuspended in 20 ml of PBS, pH 7.4, containing 1 mM EDTA, 5 mM DTT, 1 mg/ml lysozyme, and Complete Protease Inhibitor Cocktail (Roche Diagnostics, Lewes, United Kingdom). Following incubation on ice for 30 minutes, the cells were lysed in Sarkosyl (1.5% vol/vol), and cellular debris were removed by centrifugation at 300,000 g (4°C, 1 hour). Triton X-100 (2% vol/vol) was added, and the lysate was applied to a 5-ml glutathione-Sepharose column pre-equilibrated with PBS. The column was washed with 5 volumes of PBS containing 1 M NaCl, followed by 5 volumes of PBS without NaCl. Protein was eluted with 20 ml of PBS, pH 7.4, containing 20 mM reduced glutathione. Protein-containing fractions were dialyzed against Tris-buffered saline (TBS), pH 7.4, and concentrated using centrifugal ultrafiltration (Biomax-10K; Millipore Corp., Bedford, Massachusetts, USA). The IMPDH2 constructs were polyhistidine-tagged and were purified on chelating Sepharose (Pharmacia, Chalfont St. Giles, United Kingdom) charged with 50 mM CoCl2 according to the manufacturer’s instructions. The molar concentrations of proteins were determined from their absorbance at 280 nm, and extinction coefficients were calculated from the amino acid sequence. All proteins were homogeneous by SDS-PAGE, except that for the CBS1-2, CBS3-4, and CLC2 constructs there was a minor degree of proteolytic degradation (<10%). In those cases, the concentrations were corrected by densitometric estimation of the proportion of full-length protein.
Expression of AMPK heterotrimers in CCL13 cells and kinase assays.
CCL13 cells from American Type Culture Collection (Manassas, Virginia, USA) were grown in DMEM with GlutaMax-1 (GIBCO catalog no. 61965-026; Life Technologies, Paisley, United Kingdom) plus FBS (10% vol/vol). PCDNA plasmids (28–30) encoding myc-tagged α1, β1, and γ1, γ2, or γ3 were purified using HiSpeed Plasmid Maxi kits (QIAGEN, Crawley, United Kingdom), and expressed by transient transfection using SuperFect (QIAGEN Inc.) in CCL13 cells (30). After 48 hours, the cells, 80–90% of which were confluent by this time, were subjected to “rapid lysis,” and the lysates were immunoprecipitated using anti-myc antibody and assayed as described previously (31). Results are means ± SD from triplicate transfections. “Slow lysis” of CCL13 cells was as described by Stein et al. (32). AMPK activities were corrected for minor differences in expression levels by analysis of cell lysates by SDS-PAGE and probing of blots with anti–AMPK-α1 antibody (0.06 μg/ml) directly conjugated to IRDye 800 (Molecular Probes Inc., Eugene, Oregon, USA) in TBS-Tween (10 mM Tris/HCl, pH 7.4, 0.5 M NaCl, Tween-20 [1% wt/vol]) plus nonfat milk powder (1% wt/vol). After washing (six times for 5 minutes in TBS-Tween and once for 5 minutes in PBS), the remaining dye on the membrane was quantified using an Odyssey IR imager (LI-COR Biosciences, Cambridge, United Kingdom).
Ligand-binding assays.
The method to determine ligand binding was based on that of Janosik et al. (33). Various concentrations of [14C]AMP (2 MBq/μmol), [14C]S-adenosyl methionine (2 MBq/μmol), or [γ-32P]ATP (5 MBq/μmol) were incubated with glutathione-S-transferase–CBS domain fusions (1 μM for WT, up to 20 μM for some mutants) for 10 minutes at 25°C in a 20-μl total volume in TBS (50 mM Tris/HCl, pH 7.4, 150 mM NaCl). To stop the reactions, 10 μl of the reaction mixture was spotted onto a Millipore MF Filter Membrane disc (2.5 cm; Millipore Corp.), rapidly filtered under high vacuum (0.13 mbar, <1 second), and washed with 1 ml of ice-cold TBS. Radioactivity was determined on the filters by scintillation counting. Nonspecific binding (typically <200 disintegrations per minute (dpm), compared with specific binding of >4,000 dpm) was determined by control assays with glutathione-S-transferase (GST) and was subtracted. For ATP binding, 5 mM MgCl2 was routinely included; it was omitted for studies of AMP binding, although MgCl2 did not affect the binding of either nucleotide. Data were fitted to the binding models given in the text, using GraphPad Prism 3 (GraphPad Software Inc., San Diego, California, USA). Estimates of resulting parameters, ± SE, are given in the text and Table 1, and the curves shown in the figures are theoretical curves obtained using those estimates.
Table 1.
Binding parameters for AMP and ATP, measured with various constructs of AMPK, IMPDH2, and CLC2
Enzymatic assay of IMPDH2.
We assayed IMPDH2 by following the increase in absorbance at 340 nm due to formation of NADH in a BMG FLUOstar OPTIMA plate reader (BMG Lab Technologies, Aylesbury, United Kingdom) at 30°C in plate mode. The assay buffer contained 50 mM Tris/HCl, 100 mM KCl, 1 mM DTT, 3 mM EDTA, and 5 mM MgCl2. Where ATP was added, additional MgCl2 was added to maintain a constant 5-mM excess of MgCl2 over ATP. The total volume was 200 μl, and the reaction was started by addition of IMP.
Results
The N-terminal and C-terminal CBS domain pairs from AMPK-γ2 bind one molecule of AMP or ATP.
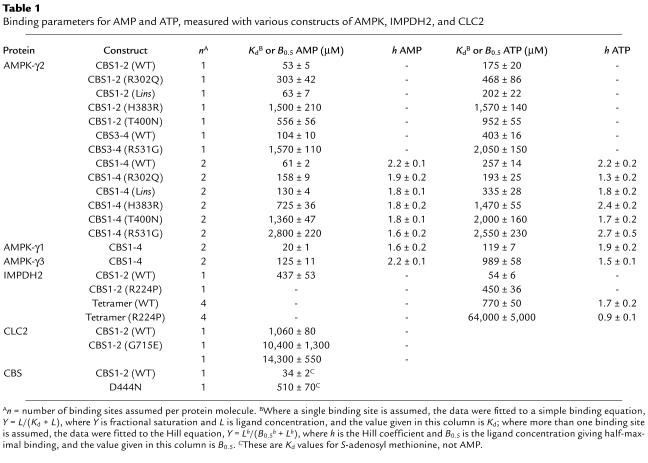
The crystal structure of a bacterial IMPDH shows that the tandem pair of CBS domains are intimately associated via hydrophobic interactions between homologous β sheets composed of three strands from one domain and one from the other (34), making it very unlikely that single domains would be stable. We therefore cloned DNA from human AMPK-γ2 encoding either all four domains (CBS1-4), or the N-terminal pair (CBS1-2) or the C-terminal pair (CBS3-4). All three constructs were expressed as GST fusions in bacteria and were purified on glutathione-Sepharose as soluble proteins of the expected sizes in good yield. Binding of AMP was assessed using [14C]AMP and a rapid-filtration method. Initial fitting of the data using the WT CBS1-2 fusion showed that the maximal binding was very close to 1 mole (0.95 ± 0.05) per mole of protein, so data were subsequently fitted to a single-site binding model (Figure 1a), yielding a dissociation constant (Kd) of 53 μM. We also used site-directed mutagenesis to create four of the mutations in CBS1-2 that cause WPWS, i.e., R302Q, H383R and T400N, and Lins (which inserts a leucine residue in the linker between CBS1 and CBS2). All mutants still bound one molecule of AMP, but the Kd values for R302Q, T400N, and H383R were increased six-, ten-, and 28-fold, respectively, relative to that for the WT (Figure 1a and Table 1). The Kd for Lins was not significantly different from that for the WT.
Figure 1.
Binding of AMP (a and c) or ATP (b and d) by the CBS1-2 construct (a and b) or the CBS3-4 construct (c and d). The fusion proteins were either the WT (open circles) or one of five point mutants (R302Q, filled circles; Lins, open squares; H383R, filled squares; T400N, open triangles; R531G, filled triangles). Data were fitted to a single-site binding model: bound = [nucleotide]/(Kd + [nucleotide]). The curves are theoretical curves obtained using the Kd values shown in Table 1.
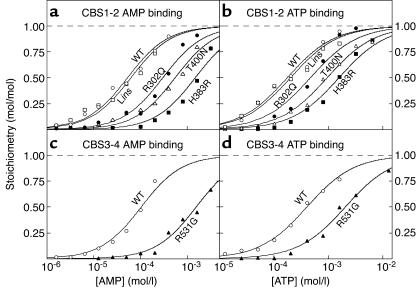
Since high concentrations of ATP antagonize activation of AMPK by AMP (35), we suspected that the CBS1-2 proteins would also bind ATP. This was indeed the case, and for the WT the data could be fitted to a single-site binding model with a Kd 3.3-fold higher than that for AMP. All of the mutants still bound ATP, but the Kd values were increased in the same rank order as for AMP binding, albeit to a lesser extent. For R302Q, T400N, and H383R, the increases were three-, five-, and ninefold, respectively, while Lins was again not significantly different from the WT (Figure 1b and Table 1). High concentrations of ATP completely displaced binding of AMP from the WT (Figure 2), showing that binding of the two nucleotides is mutually exclusive. We also measured the apparent Kd for AMP of the WT CBS1-2 protein in the presence of four different concentrations of ATP and fitted the results to the equation: apparent Kd = KdAMP(1 + [ATP]/KdATP). This yielded estimates for KdAMP and KdATP of 51 and 180 μM, respectively, very close to the estimates obtained by direct binding measurements (53 and 175 μM).
Figure 2.
Displacement of AMP from the CBS1-2 construct by ATP. A fixed concentration of [14C]AMP (180 μM) was incubated with the CBS1-2 construct in the presence of increasing concentrations of ATP. Data were fitted to the binding model: bound = AMP/[AMP + KdAMP(1 + [ATP]/KdATP)].
The CBS3-4 fusion protein bound a single molecule of AMP with a Kd of 104 μM. We also created a single point mutation in CBS3-4 that causes WPWS when present in intact γ2, i.e., R531G. The mutant still bound AMP, but the Kd increased 15-fold (Figure 1c and Table 1). The WT also bound ATP with a Kd fourfold higher than that for AMP, while the Kd for ATP was increased fivefold relative to the WT in the R531G mutant (Figure 1d and Table 1).
The four tandem CBS domains from AMPK-γ2 bind two molecules of AMP or ATP with positive cooperativity.
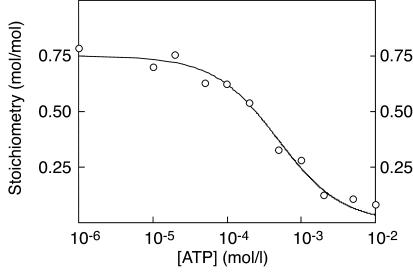
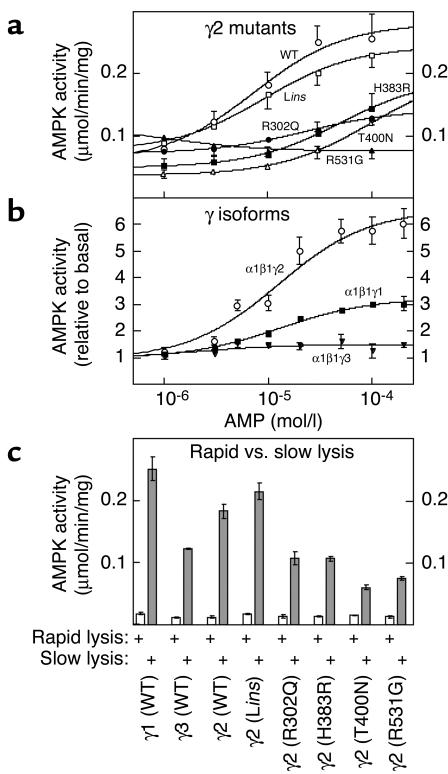
Since the CBS1-2 and the CBS3-4 fusion proteins from γ2 each bound one molecule of AMP or ATP, we expected that the CBS1-4 construct would bind two, which was indeed the case. The best fits were obtained using a Hill plot model with two identical, interacting sites: bound = 2 × [AMP]h/(B0.5h + [AMP]h), where h is the Hill coefficient and B0.5 is the concentration giving half-maximal binding (Figure 3a and Table 1). For the WT γ2 construct, this yielded a B0.5 for AMP of 61 μM. The Hill coefficient was close to 2, indicating that the two sites bind AMP with strong positive cooperativity, i.e., that once AMP is bound at the first site the affinity at the second site increases so that it fills almost immediately. Similar results were obtained for the binding of ATP (Figure 3b and Table 1), which yielded a B0.5 more than fourfold higher than that for AMP, consistent with the finding that both CBS1-2 and CBS3-4 bound ATP with lower affinity than AMP. All WPWS mutants still bound two molecules of AMP, but the B0.5 values were markedly increased (Figure 3a and Table 1). For Lins, R302Q, H383R, T400N, and R531G, the increases in B0.5 were 2.1-fold, 2.6-fold, 12-fold, 22-fold, and 46-fold, respectively. All mutants also bound two molecules of ATP, but with changes in B0.5 and/or Hill coefficient (Figure 3b and Table 1), except for the Lins mutant, which was not significantly different from the WT.
Figure 3.
(a and b) Binding of AMP (a) and ATP (b) to the GST–CBS1-4 fusion protein from γ2 and WPWS mutants. (c and d) Binding of AMP (c) and ATP (d) by the GST–CBS1-4 fusion proteins from γ1, γ2, and γ3. The methodology was as for Figure 1, except that data were fitted to a two-site Hill plot model: bound = 2 × [nucleotide]h/(B0.5h + [nucleotide]h).
Binding of AMP and ATP to the four tandem CBS domains from AMPK-γ1 and -γ3.
We also made CBS1-4 fusion proteins from the γ1 and γ3 isoforms. These also bound two molecules of AMP and ATP with positive cooperativity, but with B0.5 values that were significantly lower (γ1) or higher (γ3) than for γ2 (Table 1, Figure 3, c and d). For the constructs from γ1, γ2, and γ3, the B0.5 values for AMP were 20, 53, and 125 μm, respectively.
Effects of WPWS mutations on activation of AMPK heterotrimers by AMP.
While the results in the previous section showed that isolated CBS domain pairs bind AMP and ATP, they did not conclusively prove that these domains form the regulatory nucleotide binding sites in the heterotrimeric AMPK complex. To address this, we expressed recombinant α1β1γ2 heterotrimers in WT and mutated forms in CCL13 cells, purified them by immunoprecipitation via myc epitope tags on the α subunit, and assayed at various AMP concentrations. CCL13 cells (a human cell line now thought to be a variant of HeLa cells) were used because they have a high transfection efficiency and a low endogenous AMPK activity. Figure 4a shows that, for the Lins, R302Q, H383R, T400N, and R531G mutations, the A0.5 values (concentration causing half-maximal activation) for AMP increased in the same order in which these mutations increased the B0.5 for binding of AMP to the isolated CBS1-4 constructs, i.e., WT (7 ± 3 μM) ≈ Lins (9 ± 4 μM) < R302Q (23 ± 6 μM) < H383R (51 ± 40 μM) < T400N (95 ± 32 μM) < R531G (>100 μM). The A0.5 values for the heterotrimers were all around tenfold lower than the B0.5 values for the isolated CBS1-4 constructs, indicating that the presence of the α and/or β subunits increased the affinity for AMP. It was not possible to conduct assays above 100 μM AMP, because at these concentrations the nucleotide inhibits the activity by competing with ATP at the kinase domain on the α subunit. For this reason, the estimates of A0.5 for the H383R and T400N mutants are less accurate than the others, while no value could be obtained for the R531G mutant. The latter appeared to have a slightly elevated basal activity in the absence of AMP and was actually inhibited by addition of AMP. With some mutants, the maximal activation also appeared to be affected. For example, the R302Q mutant was only stimulated threefold by AMP, whereas the WT was stimulated sixfold.
Figure 4.
(a) Activation of recombinant α1β1γ2 heterotrimers, with or without WPWS mutations, by AMP. (b) Activation of recombinant α1β1γ1, α1β1γ2, and α1β1γ3 heterotrimers by AMP. (c) Activation of α1β1γ2 heterotrimers, with or without WPWS mutations, by slow versus rapid lysis. Plasmids expressing myc-tagged α1 and β1 plus one of the subunits γ1, γ2 (with or without WPWS mutations), and γ3 were expressed in CCL13 cells, and the recombinant complexes were immunoprecipitated using anti-myc antibodies. AMPK activity was then determined at various concentrations of AMP. In a and b, the cells were harvested by slow lysis to elicit maximal phosphorylation (32); in c, the cells were harvested by rapid or slow lysis and the assays were conducted at 200 μM AMP. Results are means ± SE for duplicate immunoprecipitations.
Effects of γ isoform on activation of AMPK heterotrimers by AMP.
Figure 4b shows that when the activations of the WT α1β1γ1, α1β1γ2, and α1β1γ3 heterotrimers were compared using the same methodology, they differed in the degree of stimulation by AMP rather than in the A0.5 for AMP. The α1β1γ1, α1β1γ2, and α1β1γ3 heterotrimers were stimulated 3.3-, 7.0-, and 1.5-fold by AMP, but the A0.5 values were similar (13 ± 2, 12 ± 3, and 2 ± 2 μM, respectively). The value of A0.5 for α1β1γ3 is approximate, because of the very small degree of stimulation by AMP with that isoform.
Mutations in γ2 do not cause constitutive activation of AMPK.
It has previously been claimed that mutations that are associated with WPWS cause constitutive activation of AMPK (12, 36). To address this, we examined the activation of expressed AMPK in CCL13 cells by slow lysis as opposed to rapid lysis. Slow lysis involves harvesting the cells by scraping them off and centrifuging them prior to resuspension in homogenization medium and activates AMPK by a combination of mechanical stress, hypoxia, and/or glucose deprivation. Rapid lysis involves pouring off the medium and lysing the cells in situ on the culture dish using ice-cold lysis buffer, and this better preserves the physiological phosphorylation status of AMPK. Figure 4c shows that the stress of slow lysis significantly activated the α1β1γ1, α1β1γ2, and α1β1γ3 heterotrimers, although the degree of activation was lowest with the α1β1γ3 complex. All of the γ2 mutants were also activated, although the degree of activation of the R302Q, H383R, T400N, and R531G mutants was significantly lower than that of the WT and the Lins mutant. There was no evidence for constitutive activation of any of the mutants when the cells were harvested by rapid lysis.
The CBS domains from IMPDH bind ATP, and this is impaired by a mutation that causes retinitis pigmentosa.
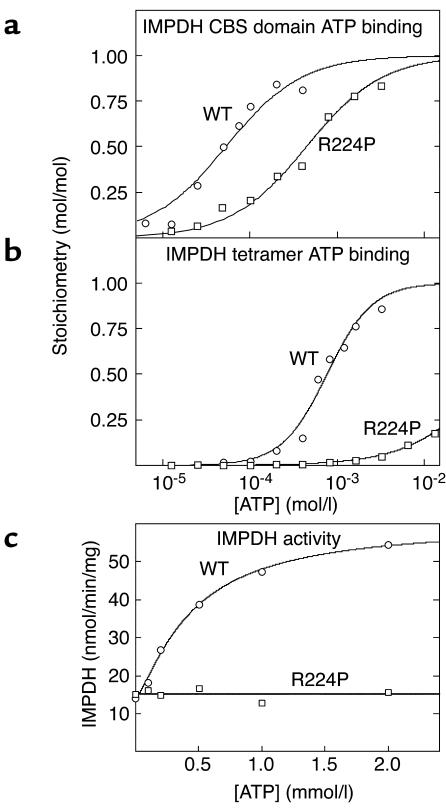
To examine whether binding of adenine nucleotides is a more general function for CBS domains, we also studied IMPDH. Retinitis pigmentosa can be caused by an R224P mutation in the second CBS domain of the IMPDH1 isoform (4). However, IMPDH2 is 84% identical in amino acid sequence with IMPDH1, and the arginine mutated in retinitis pigmentosa is conserved. Because cDNA was more readily available for IMPDH2, we cloned DNA encoding the single pair of CBS domains from that isoform, created an R224P mutation, and expressed the WT and the mutant as polyhistidine-tagged proteins in E. coli. The WT fusion protein bound one molecule of ATP with a Kd of 54 μM, while in the mutant the Kd increased more than eightfold (Figure 5a and Table 1). Using displacement of bound ATP as the assay, the WT protein also bound AMP or GMP, but only at supraphysiological concentrations (Kd = 440 and 5,760 μM, respectively).
Figure 5.
(a and b) Binding of ATP by GST fusions of the isolated CBS domain pair (residues 112–232) (a) and full-length IMPDH2 (residues 1–514) (b). (c) Activity of full-length IMPDH2 as a function of ATP concentration. Results were obtained for both the WT sequence and an R224P mutation. Data in a and b were fitted to a single-site binding model as for Figure 1. Data in c were fitted to the model: activity = basal + {[(stimulation × basal) – basal] × [ATP]h}/(A0.5h + [ATP]h).
We also cloned and expressed full-length IMPDH2 and examined the binding of ATP (Figure 5b). Interestingly, this protein (which is a tetramer) bound ATP with a B0.5 of 0.76 mM, 14-fold higher than the Kd for the isolated CBS domains, and much closer to the physiological range of ATP concentrations. An ultraviolet absorption spectrum of the purified protein showed that it did not contain any endogenous bound nucleotide. The binding of ATP to full-length IMPDH2, but not to the isolated CBS domains, also displayed positive cooperativity, with a Hill coefficient of 1.7. ATP binding by the full-length tetramer was also drastically affected by the R224P mutation, with a more than 80-fold increase in B0.5 (Figure 5b and Table 1).
ATP is an allosteric activator of IMPDH.
Since we could not find any published reports describing effects of ATP on IMPDH activity, we assayed the recombinant enzyme in the presence and absence of the nucleotide. Figure 5c shows that ATP stimulated IMPDH activity more than fourfold with a half-maximal effect at 0.44 ± 0.05 mM and a Hill coefficient of 1.3 ± 0.1. This is close to the B0.5 and Hill coefficient obtained for ATP binding to the tetramer. The effect of ATP was on Vmax, rather than on the Km for the substrates IMP or NAD+ (not shown). Even at the highest concentration used (2 mM), ATP had no effect on the R224P mutant version of the full-length tetramer (Figure 5c). This is consistent with the insignificant level of binding of ATP to the tetramer at this concentration (Figure 5b).
The CBS domain pair from the chloride channel CLC2 binds ATP, and this is severely affected by pathogenic mutations.
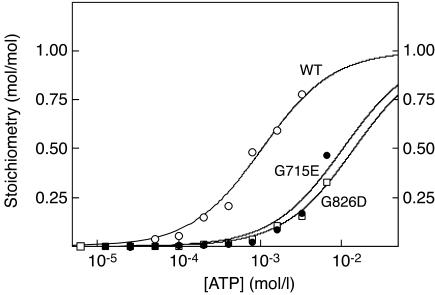
We also cloned and expressed DNA encoding the CBS domain pair from the chloride channel CLC2. Figure 6 shows that this construct bound one molecule of ATP with a Kd of 1.06 ± 0.08 mM, while G715E and G826D mutations were associated with ten- and 14-fold increases in Kd, respectively.
Figure 6.
Binding of ATP by a GST fusion of the isolated CBS domain pair (residues 582–840) from CLC2, and binding of ATP by G715E and G826D mutations. Data were fitted to a single-site binding model as for Figure 1.
The CBS domain pair from cystathionine β-synthase binds S-adenosyl methionine, and this is affected by a homocystinuria mutation.
Finally, we cloned and expressed DNA encoding the C-terminal CBS domain pair from the enzyme cystathionine β-synthase. This yielded a preparation that was homogeneous by SDS-PAGE and predominantly migrated as a monomer on native gel electrophoresis (not shown). This domain pair bound one molecule of S-adenosyl methionine (SAM) with a Kd of 34 ± 2 μM, while the D444N mutation that causes homocystinuria (2) increased the Kd for SAM 15-fold to 510 ± 70 μM (Figure 7).
Figure 7.
Binding of SAM by a GST fusion of the isolated CBS domain pair (residues 416–551) from cystathionine β-synthase, and binding of SAM by a D444N mutation. Data were fitted to a single-site binding model as for Figure 1.
Discussion
Overall, our results show that tandem pairs of CBS domains form allosteric binding sites for adenosine derivatives, i.e., AMP and ATP in the case of AMPK, ATP in the cases of IMPDH and CLC2, and SAM in the case of cystathionine β-synthase. Moreover, they show that the many mutations in CBS domains that cause human hereditary diseases invariably impair the binding of these regulatory adenosine derivatives. To our knowledge, our findings represent not only the first direct evidence as to the function of CBS domains, but also the first description of the biochemical defect in several human hereditary diseases where mutations occur in these domains.
Our results strongly support the idea that the four tandem pairs of CBS domains in the γ subunits of AMPK provide two allosteric binding sites for AMP and ATP in the heterotrimeric complex, one or both of which are therefore targets for development of drugs aimed at obesity and type 2 diabetes. The evidence in favor of this may be summarized as follows: (a) While the N-terminal and C-terminal pairs of CBS domains from the γ2 subunit both bound one molecule of AMP when expressed in and purified from bacteria, the construct containing all four domains bound two molecules of AMP. (b) Mutations that caused an increase in the B0.5 for binding of AMP to the CBS domains of γ2 also caused an increase in the A0.5 for activation of recombinant α1β1γ2 complexes, with the same order of potency, i.e., B0.5 or A0.5 for WT ≈ Lins < R302Q < H383R < T400N < R531G. (c) ATP also bound to the bacterially expressed CBS domains in a mutually exclusive manner with AMP, albeit with lower affinity. This is consistent with the fact that high concentrations of ATP inhibit allosteric activation of AMPK in a manner that appears to be competitive with AMP (35). Although mutually exclusive binding to the bacterially expressed CBS domains does not prove that AMP and ATP bind to the same site, this is the simplest explanation that is also consistent with our findings that the γ2 mutations reduced the affinity for ATP in the same order in which they reduced the affinity for AMP.
Our studies on AMP activation of heterotrimeric AMPK complexes are in good agreement with those of Daniel and Carling (37), who found that mutations associated with WPWS exhibited defective activation by AMP, with the effect on A0.5 increasing in the order WT ≈ Lins ≈ R302Q < H383R (they did not study the T400N mutant). Like we found, Daniel and Carling found that the maximal stimulation by AMP was reduced by the R302Q mutation, while with the R531G mutant the stimulation by AMP was abolished and the basal activity measured in the absence of AMP was slightly higher. They suggested that the latter might be because the inhibitory effect of ATP might also be reduced with this mutant. This is supported by our findings that the affinity of the R531G mutant for ATP, when expressed in the context of either CBS3-4 or CBS1-4 (Table 1), was greatly reduced.
Neither our results nor those of Daniel and Carling (37) support two previous claims (12, 36) that WPWS mutations make AMPK complexes constitutively active. When the cells were harvested by rapid lysis (which much better preserves the physiological phosphorylation state of AMPK), none of the mutants exhibited greater activity than the WT (Figure 4c). When the cells were harvested by the slow-lysis procedure (which causes activation of AMPK due to hypoxia, glucose deprivation, and/or mechanical stress), the mutants that exhibited defective activation by AMP in vitro (R302Q, H383R, T400N, and R531G) were all activated, although to a lower extent than the WT. Of the two previous claims that the mutations caused constitutive activation of AMPK, both relied on indirect approaches. One involved making a mutation equivalent to R302Q in the γ1 rather than the γ2 isoform (36), although naturally occurring mutations in γ1 have not been reported. The other involved making mutations equivalent to T400N and N488I in the yeast γ subunit homologue, Snf4, and analyzing two-hybrid interactions with Snf1 (the α homologue) as a surrogate measure of kinase activity (12). In fact, while both mutations did appear to cause small (twofold) increases in the Snf1/Snf4 interaction under basal conditions, removal of glucose from the medium (known to activate the SNF1 complex [ref. 38]) appeared to cause further large increases in interaction with both of the mutants as well as with the WT. The use of the term “constitutively active” to describe these Snf4 mutants (12) is therefore misleading. Our results and those of Daniel and Carling (37) suggest that while the R531G mutant has a slightly elevated basal activity (due perhaps to reduced binding of the inhibitor, ATP), the major effect of the mutations is to reduce activation of AMPK in response to stress.
Our results with IMPDH2 and CLC2 suggest that sensing of cellular energy status by binding of adenine nucleotides may be a general function of CBS domain pairs, rather than being a function restricted to AMPK. The CBS domain pairs from IMPDH2 also bound AMP and GMP in vitro in addition to ATP, but only ATP bound at physiologically relevant concentrations. IMPDHs catalyze the first step in purine nucleotide biosynthesis that is committed to synthesis of GMP rather than AMP. They are subject to feedback inhibition by GMP, but GMP did not bind with high affinity to the CBS domains. Since inhibition by GMP is competitive with the substrate, IMP (39), it is more likely that feedback inhibition is due to binding of GMP to the catalytic site. It is interesting that, while ATP bound to the isolated CBS domain pair from IMPDH2 with a Kd in the low micromolar range (54 μM) with no evidence for interaction between sites, it bound to full-length enzyme with positive cooperativity and a B0.5 (770 μM) that was 14-fold higher and much closer to the physiological range of ATP concentrations. The crystal structures of mammalian and bacterial IMPDH show that the enzyme is a tetramer with the CBS domain pairs on the outside, with subunit contacts being made entirely by the catalytic domains (34, 40). Our results suggest that the CBS domain pairs in the tetramer are constrained in a conformation that has a lower affinity for ATP than the isolated domains have, but that binding of ATP to the first site causes a conformational change that is transmitted across the subunit interface to increase the affinity of the remaining CBS domain pairs.
We were unable to find any previous reports that IMPDH was activated by ATP, but our results now show, for the first time to our knowledge, that the nucleotide increases the Vmax of the enzyme more than fourfold. The reasonably close correspondence between the B0.5 for ATP binding (770 μM) and the A0.5 for ATP activation (440 μM) of the tetramer, and the finding that the R224P mutation in the second CBS domain abolishes both binding of ATP and activation by ATP, provide strong evidence that the allosteric activation is due to binding of the nucleotide to the CBS domains. We propose that this mechanism ensures that the synthesis of guanine nucleotides only occurs when the supply of ATP is sufficient, a mechanism that accompanies feedback inhibition by GMP. This is analogous to the regulation of the key enzyme of pyrimidine nucleotide synthesis, aspartate transcarbamylase, in bacteria, which is inhibited by the end product CTP while being activated by ATP (41).
Our results also suggest that the natural ligand for the CBS domains of CLC chloride channels is ATP, and that a mutation in CLC2 (G715E) associated with idiopathic generalized epilepsy (6), and a mutation (G826D) equivalent to one in CLC1 that causes congenital myotonia (5), both lead to a severe defect in ATP binding. While our work was in progress, Vanoye and George (42) reported, using patch clamp analysis, that human CLC4 channels only supported a chloride current when incubated on the cytoplasmic side with ATP or a nonhydrolyzable analogue. The CBS domain pairs of the CLC family are predicted to lie on the cytoplasmic side of the membrane. Our results and those of Vanoye and George suggest that binding of ATP to the CBS domains of the CLC chloride channels is necessary before the channels will open.
We therefore propose that, in most cases, tandem pairs of CBS domains form sensors of cellular energy status that act by binding AMP and/or ATP. An exception to this appears to be cystathionine β-synthase itself, which catalyzes the first step in cysteine synthesis and is allosterically activated by SAM. Its substrate, homocysteine, is an intermediate in the “activated methyl cycle” in which SAM (an important donor of methyl groups during biosynthesis) is regenerated from S-adenosyl homocysteine. A low activity of cystathionine β-synthase would promote recycling of homocysteine into SAM, whereas activation of the enzyme by high concentrations of SAM would favor removal of homocysteine from the cycle and its conversion to cysteine instead. Intriguingly, point mutations in the CBS domains of cystathionine β-synthase, or premature termination or proteolysis that removes them, result in enzyme that no longer responds to SAM (2, 43, 44). Kraus and coworkers (33) provided evidence that the CBS domains form an autoinhibitory domain whose effect is relieved by SAM binding, which is consistent with the idea that the CBS domains might represent the binding site for SAM, although this had not been directly demonstrated. Our present results now provide strong support for this hypothesis, since the isolated CBS domains bound one molecule of SAM with a Kd of 34 μM, while the D444N mutation increased this to approximately 500 μM (Figure 7). These results are consistent with previous estimates of SAM binding to the full-length enzyme of around 15 μM (33), and with results showing that WT cystathionine β-synthase was activated by SAM with a half-maximal effect at 7 μM, while in the D444N mutant this was increased to 460 μM (45). The pathogenic effect of the D444N mutation might also be due in part to a reduced expression of the protein (45).
In the case of cystathionine β-synthase, the CBS domains appear to have been adapted to bind not an adenine nucleotide, but a related adenosine-containing compound, SAM. In fact the second CBS domain in cystathionine β-synthase is rather poorly conserved (ref. 1; see also the PFAM database, entry no. PF00571, ref. 46). In addition, a truncation that removes the N-terminal 70 residues of cystathionine β-synthase, which contains the heme-binding region, results in enzyme that retains 20% of WT activity but is no longer activated by SAM (47). One possibility is that the CBS domains of cystathionine β-synthase bind the adenosyl moiety of SAM, whereas the N-terminal heme domain is involved in binding of the methionine moiety, which is not present in ligands that bind to other CBS domain pairs.
Although our present results provide strong support for the idea that the CBS domains of the γ subunits of AMPK provide the allosteric binding sites for AMP and ATP, a number of puzzling findings remain to be addressed in future studies. Firstly, the A0.5 values for activation of WT and mutant versions of the α1β1γ2 complex by AMP were generally around tenfold lower than the B0.5 values for binding of AMP to the equivalent CBS1-4 constructs. Secondly, the B0.5 for binding of AMP to the WT CBS1-4 constructs was different for the three γ subunit isoforms (γ1, 20 μM; γ2, 53 μM; γ3, 125 μM), although the recombinant α1β1γ1, α1β1γ2, and α1β1γ3 complexes were all activated by similar, lower concentrations of AMP (A0.5 = 2–13 μM) and differed instead in their degree of stimulation by AMP (Figure 4b), in agreement with previous results obtained with native complexes prepared by immunoprecipitation (28). Another unexpected finding was that the CBS domains of γ2 contain two binding sites for both AMP and ATP. There is an interesting parallel here with the regulatory subunits of cAMP-dependent protein kinase, which contain two tandem binding sites for cAMP, although these are not related to CBS domains. A current model for activation of cAMP-dependent protein kinase is that cAMP binds initially to the C-terminal site (site B), and only then does site A become accessible, although it is binding to site A that causes activation (48). Further work is required to determine whether a similar mechanism operates in the case of AMPK. The finding that mutations in CBS1 (R302Q), CBS2 (H383R and T400N), and CBS4 (R531G) all affect AMP activation (Figure 4a) suggests that AMP must bind to both sites for activation to occur. However, a sigmoidal activation of AMPK by AMP (which might be expected if occupancy of both binding sites were necessary for activation) has not been reported to our knowledge.
Another puzzling finding is that, although the Saccharomyces cerevisiae homologues of the AMPK γ subunits (Snf4p) and cystathionine β-synthase both contain CBS domains in the same position as their human counterparts, they have not been found to be activated by AMP (38) or SAM (49, 50), respectively.
Finally, one interesting feature of the pathogenic mutations in CBS domains is that they tend to occur in equivalent positions. Thus, the R302Q, H383R, and R531G mutations in CBS1, CBS2, and CBS4 of γ2 all align (plus or minus one residue) with the D444N mutation in cystathionine β-synthase and with the R200Q mutation in CBS1 of the γ3 isoform of pig AMPK (13). Similarly, the T400N mutation in CBS2 of γ2 aligns (plus or minus one residue) with the R224P mutation in CBS2 of IMPDH1. These “hot spots” for pathogenic mutations are likely to be directly involved in binding of the adenosine-containing ligand. A model for the N-terminal domain pair in the γ2 subunit of AMPK, based on the atomic coordinates of a bacterial IMPDH (34), is shown in Figure 8. Two four-stranded β sheets form a deep hydrophobic cleft between the two domains that is of suitable dimensions to accommodate an adenosine moiety. Many of the residues that are mutated in disease states have basic, positively charged side chains (e.g., Arg302 and His383 in γ2, and Arg224 in IMPDH2, which aligns with His401 in γ2) and are predicted to lie around the mouth of this cleft (Figure 8). In the CBS domain pairs that bind AMP and/or ATP, these residues form a positively charged patch at the mouth of the cleft, which is likely to bind the α phosphate moiety of adenine nucleotides. Other residues that are mutated (Thr400 in γ2, and Gly828 in CLC2, which aligns with Tyr397 in γ2) are predicted to lie within the cleft itself, where they may form interactions with the ribose or adenine moieties. Another notable feature is that the basic residues that form the basic, positively charged patch at the mouth of the cleft in γ2 are either uncharged or acidic in cystathionine β-synthase. The D444N mutation in the latter protein would neutralize a negative charge that occurs in the equivalent position to Arg302 in γ2. Intriguingly, the ligand for cystathionine β-synthase, i.e., SAM, has a positively charged sulfur atom in approximately the same position as the negatively charged phosphate of AMP, and it is likely that Asp444 forms an electrostatic interaction with this sulfur atom.
Figure 8.
Model of the N-terminal pair of CBS domains (CBS1 and CBS2) from the γ2 subunit of AMPK. The picture is a view of a molecular-surface representation made using the program GRASP (Department of Biochemistry, Columbia University, New York, New York, USA) (51), with CBS1 on the left. The model was made using MODELLER 6 (Department of Biopharmaceutical Sciences, University of California San Francisco, San Francisco, California, USA) (52) and was based on the atomic coordinates of the CBS domain pair from a bacterial IMPDH (Protein Data Bank code 1ZFJ) (34). Electrostatic potential at the surface is depicted as red for negative and blue for positive. The approximate positions of residues mutated in WPWS are shown. R302 and H383 occupy equivalent positions in CBS1 and CBS2, respectively, and are adjacent to each other in the model. Part of residue T400 projects into the hydrophobic cleft between the two domains.
Acknowledgments
This study was supported by Research and Technological Development contract QLG1-CT-2001-01488 from the European Commission and a Programme Grant from the Wellcome Trust. M. Anis was supported by a studentship from the Biotechnology and Biological Sciences Research Council (United Kingdom). We thank Fahriea Anis, David Liu, Marcus Lyall, and Andrew Ferenbach for help with cloning of DNA constructs, and David Carling for the original plasmids encoding AMPK subunits.
Footnotes
See the related Commentary beginning on page 182.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Wolff-Parkinson-White syndrome (WPWS); AMP-activated protein kinase (AMPK); IMP dehydrogenase (IMPDH); Tris-buffered saline (TBS); disintegrations per minute (dpm); glutathione-S-transferase (GST); Hill coefficient (h); concentration causing half-maximal binding (B0.5); concentration causing half-maximal activation (A0.5); S-adenosyl methionine (SAM).
References
- 1.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 2.Kluijtmans LA, et al. Defective cystathionine β-synthase regulation by S-adenosylmethionine in a partially pyridoxine responsive homocystinuria patient. J. Clin. Invest. 1996;98:285–289. doi: 10.1172/JCI118791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowne SJ, et al. Mutations in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) cause the RP10 form of autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2002;11:559–568. doi: 10.1093/hmg/11.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennan A, et al. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild-type and Rho(–/–) mice. Hum. Mol. Genet. 2002;11:547–557. doi: 10.1093/hmg/11.5.547. [DOI] [PubMed] [Google Scholar]
- 5.Pusch M. Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum. Mutat. 2002;19:423–434. doi: 10.1002/humu.10063. [DOI] [PubMed] [Google Scholar]
- 6.Haug K, et al. Mutations in CLCN2 encoding a voltage-gated chloride channel are associated with idiopathic generalized epilepsies. Nat. Genet. 2003;33:527–532. doi: 10.1038/ng1121. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd SE, et al. Characterisation of renal chloride channel, CLCN5, mutations in hypercalciuric nephrolithiasis (kidney stones) disorders. Hum. Mol. Genet. 1997;6:1233–1239. doi: 10.1093/hmg/6.8.1233. [DOI] [PubMed] [Google Scholar]
- 8.Konrad M, et al. Mutations in the chloride channel gene CLCNKB as a cause of classic Bartter syndrome. J. Am. Soc. Nephrol. 2000;11:1449–1459. doi: 10.1681/ASN.V1181449. [DOI] [PubMed] [Google Scholar]
- 9.Gollob MH, et al. Novel PRKAG2 mutation responsible for the genetic syndrome of ventricular preexcitation and conduction system disease with childhood onset and absence of cardiac hypertrophy. Circulation. 2001;104:3030–3033. doi: 10.1161/hc5001.102111. [DOI] [PubMed] [Google Scholar]
- 10.Gollob MH, et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N. Engl. J. Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 11.Blair E, et al. Mutations in the gamma-2 subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum. Mol. Genet. 2001;10:1215–1220. doi: 10.1093/hmg/10.11.1215. [DOI] [PubMed] [Google Scholar]
- 12.Arad M, et al. Constitutively active AMP kinase mutations cause glycogen storage disease mimicking hypertrophic cardiomyopathy. J. Clin. Invest. 2002;109:357–362. doi:10.1172/JCI200214571. doi: 10.1172/JCI14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milan D, et al. A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 2000;288:1248–1251. doi: 10.1126/science.288.5469.1248. [DOI] [PubMed] [Google Scholar]
- 14.Hardie DG, Hawley SA. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays. 2001;23:1112–1119. doi: 10.1002/bies.10009. [DOI] [PubMed] [Google Scholar]
- 15.Song XM, et al. 5-Aminoimidazole-4-carboxamide ribonucleoside treatment improves glucose homeostasis in insulin-resistant diabetic (ob/ob) mice. Diabetologia. 2002;45:56–65. doi: 10.1007/s125-002-8245-8. [DOI] [PubMed] [Google Scholar]
- 16.Buhl ES, et al. Long-term AICAR administration reduces metabolic disturbances and lowers blood pressure in rats displaying features of the insulin resistance syndrome. Diabetes. 2002;51:2199–2206. doi: 10.2337/diabetes.51.7.2199. [DOI] [PubMed] [Google Scholar]
- 17.Iglesias MA, et al. AICAR administration causes an apparent enhancement of muscle and liver insulin action in insulin-resistant high-fat-fed rats. Diabetes. 2002;51:2886–2894. doi: 10.2337/diabetes.51.10.2886. [DOI] [PubMed] [Google Scholar]
- 18.Bergeron R, et al. Effect of 5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside infusion on in vivo glucose and lipid metabolism in lean and obese Zucker rats. Diabetes. 2001;50:1076–1082. doi: 10.2337/diabetes.50.5.1076. [DOI] [PubMed] [Google Scholar]
- 19.Winder WW, Hardie DG. Inactivation of acetyl-CoA carboxylase and activation of AMP-activated protein kinase in muscle during exercise. Am. J. Physiol. 1996;270:E299–E304. doi: 10.1152/ajpendo.1996.270.2.E299. [DOI] [PubMed] [Google Scholar]
- 20.Merrill GM, Kurth E, Hardie DG, Winder WW. AICAR decreases malonyl-CoA and increases fatty acid oxidation in skeletal muscle of the rat. Am. J. Physiol. 1997;273:E1107–E1112. doi: 10.1152/ajpendo.1997.273.6.E1107. [DOI] [PubMed] [Google Scholar]
- 21.Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 2001;108:1167–1174. doi:10.1172/JCI200113505. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawley SA, Gadalla AE, Olsen GS, Hardie DG. The anti-diabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes. 2002;51:2420–2425. doi: 10.2337/diabetes.51.8.2420. [DOI] [PubMed] [Google Scholar]
- 23.Fryer LG, Parbu-Patel A, Carling D. The anti-diabetic drugs rosiglitazone and metformin stimulate AMP-activated protein kinase through distinct pathways. J. Biol. Chem. 2002;277:25226–25232. doi: 10.1074/jbc.M202489200. [DOI] [PubMed] [Google Scholar]
- 24.Minokoshi Y, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415:339–343. doi: 10.1038/415339a. [DOI] [PubMed] [Google Scholar]
- 25.Yamauchi T, et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 2002;6:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 26.Winder WW, Hardie DG. AMP-activated protein kinase, a metabolic master switch: possible roles in type 2 diabetes. Am. J. Physiol. 1999;277:E1–E10. doi: 10.1152/ajpendo.1999.277.1.E1. [DOI] [PubMed] [Google Scholar]
- 27.Moller DE. New drug targets for type 2 diabetes and the metabolic syndrome. Nature. 2001;414:821–827. doi: 10.1038/414821a. [DOI] [PubMed] [Google Scholar]
- 28.Cheung PCF, Salt IP, Davies SP, Hardie DG, Carling D. Characterization of AMP-activated protein kinase γ subunit isoforms and their role in AMP binding. Biochem. J. 2000;346:659–669. [PMC free article] [PubMed] [Google Scholar]
- 29.Woods A, et al. Characterization of AMP-activated protein kinase β and γ subunits: assembly of the heterotrimeric complex in vitro. J. Biol. Chem. 1996;271:10282–10290. doi: 10.1074/jbc.271.17.10282. [DOI] [PubMed] [Google Scholar]
- 30.Woods A, Salt I, Scott J, Hardie DG, Carling D. The α1 and α2 isoforms of the AMP-activated protein kinase have similar activities in rat liver but exhibit differences in substrate specificity in vitro. FEBS Lett. 1996;397:347–351. doi: 10.1016/s0014-5793(96)01209-4. [DOI] [PubMed] [Google Scholar]
- 31.Hardie DG, Salt IP, Davies SP. Analysis of the role of the AMP-activated protein kinase in the response to cellular stress. Methods Mol. Biol. 2000;99:63–75. doi: 10.1385/1-59259-054-3:63. [DOI] [PubMed] [Google Scholar]
- 32.Stein SC, Woods A, Jones NA, Davison MD, Carling D. The regulation of AMP-activated protein kinase by phosphorylation. Biochem. J. 2000;345:437–443. [PMC free article] [PubMed] [Google Scholar]
- 33.Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Regulation of human cystathionine beta-synthase by S-adenosyl-L-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry. 2001;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- 34.Zhang R, et al. Characteristics and crystal structure of bacterial inosine-5′-monophosphate dehydrogenase. Biochemistry. 1999;38:4691–4700. doi: 10.1021/bi982858v. [DOI] [PubMed] [Google Scholar]
- 35.Corton JM, Gillespie JG, Hawley SA, Hardie DG. 5-Aminoimidazole-4-carboxamide ribonucleoside: a specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton SR, et al. An activating mutation in the γ1 subunit of the AMP-activated protein kinase. FEBS Lett. 2001;500:163–168. doi: 10.1016/s0014-5793(01)02602-3. [DOI] [PubMed] [Google Scholar]
- 37.Daniel TD, Carling D. Functional analysis of mutations in the γ2 subunit of AMP-activated protein kinase associated with cardiac hypertrophy and Wolff-Parkinson-White syndrome. J. Biol. Chem. 2002;277:51017–51024. doi: 10.1074/jbc.M207093200. [DOI] [PubMed] [Google Scholar]
- 38.Wilson WA, Hawley SA, Hardie DG. The mechanism of glucose repression/derepression in yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert HJ, Lowe CR, Drabble WT. Inosine 5′-monophosphate dehydrogenase of Escherichia coli. Purification by affinity chromatography, subunit structure and inhibition by guanosine 5′-monophosphate. Biochem. J. 1979;183:481–494. doi: 10.1042/bj1830481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sintchak MD, et al. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell. 1996;85:921–930. doi: 10.1016/s0092-8674(00)81275-1. [DOI] [PubMed] [Google Scholar]
- 41.Fetler L, Vachette P. The allosteric activator Mg-ATP modifies the quaternary structure of the R-state of Escherichia coli aspartate transcarbamylase without altering the T<—>R equilibrium. J. Mol. Biol. 2001;309:817–832. doi: 10.1006/jmbi.2001.4681. [DOI] [PubMed] [Google Scholar]
- 42.Vanoye CG, George AG., Jr Functional characterization of recombinant human ClC-4 chloride channels in cultured mammalian cells. J. Physiol. 2002;539:373–383. doi: 10.1113/jphysiol.2001.013115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan X, Dunbrack RL, Jr, Christopher SA, Kruger WD. Mutations in the regulatory domain of cystathionine β-synthase can functionally suppress patient-derived mutations in cis. Hum. Mol. Genet. 2001;10:635–643. doi: 10.1093/hmg/10.6.635. [DOI] [PubMed] [Google Scholar]
- 44.Kery V, Poneleit L, Kraus JP. Trypsin cleavage of human cystathionine β-synthase into an evolutionarily conserved active core: structural and functional consequences. Arch. Biochem. Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 45.Evande R, Blom H, Boers GH, Banerjee R. Alleviation of intrasteric inhibition by the pathogenic activation domain mutation, D444N, in human cystathionine beta-synthase. Biochemistry. 2002;41:11832–11837. doi: 10.1021/bi026248d. [DOI] [PubMed] [Google Scholar]
- 46.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveriusova J, Kery V, Maclean KN, Kraus JP. Deletion mutagenesis of human cystathionine beta-synthase. Impact on activity, oligomeric status, and S-adenosylmethionine regulation. J. Biol. Chem. 2002;277:48386–48394. doi: 10.1074/jbc.M207087200. [DOI] [PubMed] [Google Scholar]
- 48.Su Y, et al. Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains. Science. 1995;269:807–813. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- 49.Jhee KH, McPhie P, Miles EW. Yeast cystathionine beta-synthase is a pyridoxal phosphate enzyme but, unlike the human enzyme, is not a heme protein. J. Biol. Chem. 2000;275:11541–11544. doi: 10.1074/jbc.c000056200. [DOI] [PubMed] [Google Scholar]
- 50.Maclean KN, Janosik M, Oliveriusova J, Kery V, Kraus JP. Transsulfuration in Saccharomyces cerevisiae is not dependent on heme: purification and characterization of recombinant yeast cystathionine beta-synthase. J. Inorg. Biochem. 2000;81:161–171. doi: 10.1016/s0162-0134(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 51.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 52.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]