Abstract
The Drosophila homolog of the retinoid X receptor, ultraspiracle (USP), heterodimerizes with the ecdysone receptor (EcR) to form a functional complex that mediates the effects of the steroid molting hormone ecdysone by activating and repressing expression of ecdysone response genes. As with other retinoid X receptor heterodimers, EcR/USP affects gene transcription in a ligand-modulated manner. We used in vivo, cell culture, and biochemical approaches to analyze the functions of two usp alleles, usp3 and usp4, which encode stable proteins with defective DNA-binding domains. We observed that USP is able to activate as well as repress the Z1 isoform of the ecdysone-responsive broad complex (BrC-Z1). Activation of BrC-Z1 as well as EcR, itself an ecdysone response gene, can be mediated by both the USP3 and USP4 mutant proteins. USP3 and USP4 also activate an ecdysone-responsive element, hsp27EcRE, in cultured cells. These results differ from the protein null allele, usp2, which is unable to mediate activation [Schubiger, M. & Truman, J. W. (2000) Development 127, 1151–1159]. BrC-Z1 repression is compromised in all three usp alleles, suggesting that repression involves the association of USP with DNA. Our results distinguish two mechanisms by which USP modulates the properties of EcR: one that involves the USP DNA-binding domain and one that can be achieved solely through the ligand-binding domain. These newly revealed properties of USP might implicate similar properties for retinoid X receptor.
The nuclear receptors comprise a large family of transcriptional regulators that are structurally related in two evolutionarily conserved regions: a core DNA-binding domain (DBD) and a carboxyl-terminal ligand-binding domain (LBD) (1). Many nuclear receptors function as ligand-regulated transcription factors, including the retinoic acid receptor and the thyroid hormone receptor, whose transcriptional properties require dimerization with a second nuclear receptor, the retinoid X receptor (RXR). Both RXR and its partner recognize and bind target DNA sequences (2–5). In cell culture, RXR heterodimers repress transcription in the absence of ligand by nucleating repressive complexes involving histone deacetylase activity (6–9). In the presence of ligand, corepressors are unable to bind the nuclear receptor complex, which remains bound to target DNA, and a coactivator complex is formed containing histone acetyltransferase activity (8, 9). The chromatin structure becomes accessible for general transcriptional machinery and results in gene activation. Ligand-regulated properties of the RXR heterodimers are conserved in invertebrates. The functional ecdysone receptor is composed of a heterodimer between the ecdysone-binding ecdysone receptor (EcR) and the RXR homologue ultraspiracle (USP) (10–13). EcR/USP complexes repress transcription in the absence of ligand and recruit coactivators in the presence of ligand (14–16).
The steroid hormone ecdysone, and its more active form 20-hydroxyecdysone, plays a crucial role in larval and metamorphic molts, and the hierarchy of genes responding to ecdysone at these stages has been well characterized and includes EcR itself (17, 18). Because of the functional conservation between vertebrate and invertebrate RXR heterodimers in cell culture, we can use the easily manipulated and well-studied Drosophila system to dissect the functional abilities of these complexes in vivo and in vitro. Previous studies have investigated the effects of disrupting the function of usp by looking at usp mutant patches of tissue in a heterozygous animal. These experiments have found a repressive role for usp in Drosophila development, while confirming the role of usp as a mediator of the ecdysone response in vivo (19, 20).
We extend these studies by analyzing in vivo the functional abilities of different usp alleles to activate and repress the ecdysone response genes EcR and the Z1 isoform of broad complex (BrC-Z1) (18, 21–22). Biochemical and cell culture data have been combined with these experiments to suggest that the USP DNA-binding domain is dispensable for the activation but not repression of certain ecdysone response targets. Given the conservation in regulatory and transcriptional properties between EcR/USP and vertebrate RXR heterodimers, the information gained from these experiments may shed new light on the roles of RXR in mediating retinoic acid and thyroid hormone signaling.
Materials and Methods
Drosophila Stocks and Transgenic Fly Lines.
The generation and detection of usp3 and usp4 mitotic clones in imaginal discs and salivary glands was carried out by using the FRT-Flp system (19, 23). Males of the genotype w1118 usp3 (f P{ry+t7.2 = neoFRT}18A); λ10 Tb/TM3 or y1w1118 usp4 (P{ry+t7.2 = neoFRT}18A); λ10 Tb/TM3 were crossed to P{w+mC = piM}10D M(1)Osp P{ry+t7.2 = neoFRT}18A/FM7; hsFLP38/hsFLP38 or P{w+mC = NM}8A M(1)OSp P{ry+t7.2 = neoFRT}18A/FM7; hsFLP38/hsFLP38 females. λ10 is an 8-kb usp+ genomic transgene inserted on the third chromosome (24). To generate imaginal disk clones, progeny were heat-shocked during the first to second instar (24–48 h old) for 1 h at 37°C to induce mitotic recombination at the FRT sites via expression of the flipase (Flp). To generate salivary gland clones, progeny were heat-shocked when 5–24 h old. Tb+ larvae were dissected during the third larval instar. Homozygous usp+ clones did not survive because of the presence of a recessive lethal M(1)Osp on the usp+ chromosome. usp2 clones were generated by crossing usp2 P{ry+t7.2 = neoFRT}18A/FM7 females with w1118 P{w+mC = piM}5A P{w+mC = piM}10D P{ry+t7.2 = neoFRT}18A; hsFLP38/hsFLP38 males. Imaginal disk clones were generated as described. As the usp+ chromosome in these experiments does not contain a recessive lethal, both usp+/usp+ and usp2/usp2 clones were created.
For the rescue of usp clones, fly stocks were generated containing w1118 usp3 UAS-usp+ f P{ry+t7.2 = neoFRT}18A; λ10 Tb/TM3. Females of this genotype were crossed to hairyIJ3-Gal4 males (25). Males from this cross of the genotype w1118 usp3 UAS-usp f P{ry+t7.2 = neoFRT}18A; λ10 Tb/hairyIJ3-Gal4 were crossed to P{w+mC = piM}10D M(1)OSp P{ry+t7.2 = neoFRT}18A/FM7; hsFLP38/hsFLP38 or P{w+mC = NM}10D M(1)OSp P{ry+t7.2 = neoFRT}18A/FM7; hsFLP38/hsFLP38 females and imaginal disc clones were generated as above.
Immunodetection.
Fixing and staining procedures have been described (23). To stain for EcR, several mouse monoclonal antibodies were used. 15C3 and 10Fil, Manduca sexta EcR antibodies able to recognize all Drosophila EcR isoforms, were used on imaginal discs at a 1:100 dilution each. 15C3 and 10Fil were developed by Dr. L. Riddiford (26) and obtained from the Developmental Studies Hybridoma Bank. Salivary glands were stained with 15C3 or AD4.4 (gift from D. Hogness; ref. 27), which recognizes EcRB1 at a 1:100 or 1:50 dilution, respectively. To stain for broad complex (BrC), two mouse antibodies were used (gifts from G. Guild, University of Pennsylvania), a Z1 isoform (3C11.OA1) and one that recognizes all BrC isoforms (Mab 2539) (21). Mab 2539 was used diluted 1:100; 3C11.OA1 was used at a dilution of 1:100. A monoclonal mouse USP antibody AB11 (gift from F. Kafatos, ref. 28) was used at a 1:100 dilution. Secondary antibodies used were Texas red-conjugated goat anti-rabbit and FITC conjugated goat anti-mouse (The Jackson Laboratory) diluted 1:100. Confocal images were collected on a Nikon/Bio-Rad confocal microscope.
Plasmids.
Cloning the usp+ cDNA into the EcoRI site of pUAST generated UAS-usp+. DNA was injected into w1118 embryos to generate UAS-usp fly lines. The reporter plasmids containing tk-hsp27EcRE, tk-DR1x2, tk-DR1x3, and pMH100-tk-luc, as well as cytomegalovirus promoter-driven expression plasmids (pCMX) expressing USP, USP3, USP4, VP16:USP, EcRB1, and β-galactosidase were described (12, 13, 16, 29, 30). pCMX-based plasmids corresponding to USPL, RXRL, VP16:USP3, VP16:USP4, VP16:USPD, Gal4:EcRB1, or His-USP were constructed by using standard techniques, including various enzyme digestions or PCR amplification. Detailed information is available on request.
Gel Mobility Shifts.
Gel mobility shift experiments were performed by mixing either bacterially expressed His-tagged USP or in vitro translated USP proteins with the appropriate DNA in 20 μl of binding solution, containing 10 mM Tris-Cl (pH 7.5)/0.05 mM EDTA/40 mM NaCl/20 mM KCl/5% glycerol/0.05% Nonidet P-40/0.2 nmol MgCl2/1 mg BSA/2 mg of poly dI:dC. The mix was incubated at room temperature for 20 min before being loaded onto a 5% polyacrylamide gel, which ran at 120–180 volts in 0.5 × TBE. For the supershift experiment described in Fig. 5A, the USP antibody AB11 was used (28).
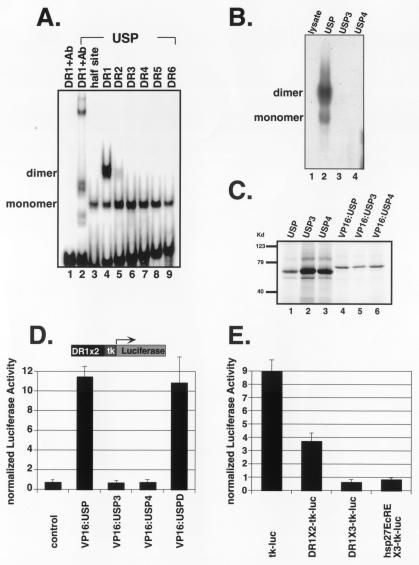
Figure 5.
The USP homodimer/DR1 complex is disrupted by USP3 and by USP4. (A) Gel mobility shift experiments in which 32P-labeled serial DR elements were incubated with bacterially expressed His-tagged USP. USP binds as a monomer on a half site, as well as on all DR elements (lanes 3–9); the USP homodimer specifically binds DR1 (lane 4). A USP specific antibody (AB11) was used in lanes 1 and 2. (B) Gel mobility shift in which a 32P-labeled DR1 element was incubated with mock translation mix (lane 1), in vitro translated USP (lane 2), USP3 (lane 3), or USP4 (lane 4). (C) In vitro translated 35S-labeled proteins corresponding to USP and its variants. (D) Transient transfection in CV-1 cells with a reporter, DR1 × 2-tk-luc, and pCMX-based plasmids expressing VP16:USP (lane 2), VP16:USP3 (lane 3), VP16:USP4 (lane 4), or VP16:USPD (lane 5). USPD represents the DNA-binding domain of USP (amino acids 50–205). In this and other experiments, luciferase activities were normalized by cotransfecting cells with CMX-lacZ, which expresses β-galactosidase. (E) Drosophila Schneider S2 cells were transfected with tk-luc (lane 1), DR1X2-tk-luc (lane 2), DR1X3-tk-luc (lane 3), and hsp27EcREX3-tk-luc (lane 4).
Cell Culture and Transfection.
Cell culture and transfection procedures for CV-1 cells were described previously (16). For transfecting Drosophila Schneider 2 cells, the calcium phosphate precipitation method was used. All experiments were performed in triplicate and repeated with similar results.
Results
usp Alleles Differ in Activating Abilities.
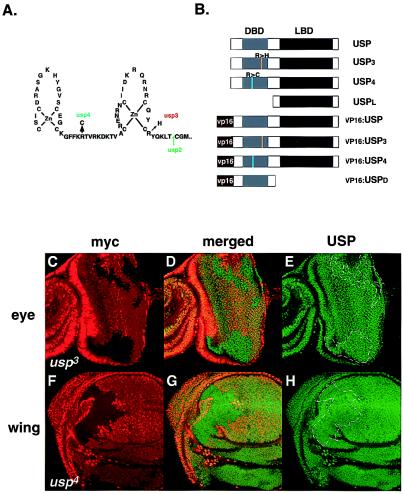
In Drosophila, we are able to generate mitotic clones mutant for usp in larval or adult tissue and use these to study the role of usp in the ecdysone cascade. Previous experiments have provided evidence that functional usp is required for the activation of EcR and repression of the Z1 isoform of the broad complex (BrC-Z1), both primary ecdysone response genes (20). As the EcR activation result was obtained by using the protein null usp2 allele, we further examined the responses of EcR and BrC-Z1 with the usp3 and usp4 alleles. These alleles are point mutations in the DNA-binding domain and produce proteins with defects in DNA binding (31) (Fig. 1A). In vivo, both usp3 and usp4 mutant clones contain wild-type levels of these mutant proteins (Fig. 1 C–H).
Figure 1.
usp alleles, expression levels, and USP variants for cell culture experiments. (A) Mapped usp mutations (31). (B) Diagram of different usp constructs, representing USP and its variants used in transfection experiments. USPL has the amino acids 1–205 of USP removed and contains the ligand-binding domain only. Placing amino acids 50–508 of wild-type or mutant USP C-terminal to the viral VP16 transactivating domain generated VP16:USP fusions. VP16:USPD is a VP16 fusion containing the DNA-binding domain (amino acids 50–205) of USP only. (C–H) Clonal regions (marked with dotted lines) of usp3 in an eye imaginal disc (C–E) and of usp4 in a wing imaginal disc (F–H) isolated from third instar wandering stage larvae and stained with an anti-myc antibody (Texas red) and with an anti-USP antibody (FITC). Clonal regions have lost the myc marker. The eye disc is situated with its differentiated cells, which are posterior to the morphogenetic furrow, facing to the right; the wing disc is situated with its anterior to the left, ventral to the top. The center panels are a merge of the Texas red and FITC pictures. Magnification is at 40×.
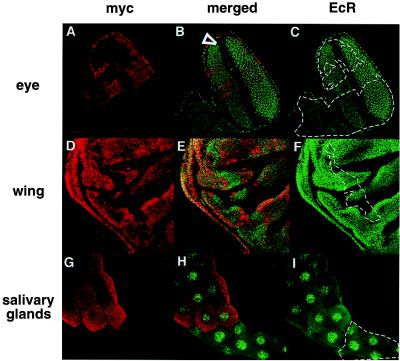
In usp2 mitotic clones, ecdysone-induced expression of EcR is missing in imaginal discs (20). However, EcR expression is unaffected in usp3 clones in both imaginal tissues and salivary glands (Fig. 2). Similar results are seen for usp4 (data not shown). The differences in the response of EcR to usp2 and to usp3 or usp4 show that USP function is necessary for ecdysone induction of EcR and that the function of the USP DBD may not be necessary for the activation of some targets.
Figure 2.
EcR expression is not altered in usp3 clones. usp3 clones (dotted lines) in an eye disc (A–C), in a wing disc (D–F), and in a salivary gland (G–I) isolated from third instar wandering larvae and stained with anti-myc antibodies (Texas red) and with anti-EcR antibodies (FITC). The eye is situated with its anterior to the bottom left. The morphogenetic furrow is marked with an arrowhead (A–C). The wing disc is situated anterior to the left, ventral to the top (D–F). Magnification for all tissues is 40×.
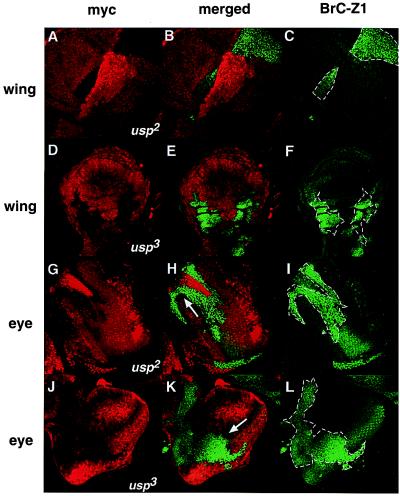
Despite the clear difference between usp3, usp4, and usp2 in EcR activation, all three alleles fail to repress BrC-Z1 (20) (Fig. 3). Using all three alleles, BrC-Z1 is up-regulated in wing imaginal discs during the late third instar stage, when expression of BrC-Z1 is normally low (Fig. 3 A–F). In the eye imaginal disc, BrC-Z1 is normally expressed posterior to the morphogenetic furrow (Fig. 3 H-I, K-L). However, in usp2 or usp3 clones, BrC-Z1 expression occurs anterior to the furrow, where it is normally repressed (Fig. 3 G–L). These results in both the wing and the eye discs indicate that the DNA-binding function of USP is necessary for repression of an ecdysone-responsive gene.
Figure 3.
BrC-Z1 is up-regulated in both usp2 and usp3 clones. Clonal studies for usp2 in a wing disk (A–C) and in an eye disc (G–I); clonal studies for usp3 in a wing disc (D–F) and in an eye disc (J–L) stained with anti-myc (Texas red) and with anti-BrC-Z1 antibodies (FITC). (A–C) The wing disk is situated anterior to the upper left, ventral to the upper right. Magnification is at 40×. (D–F) Anterior is to the left, ventral to the top. Magnification is at 20×. (G–I) The eye disc has its anterior to the bottom left. Magnification is 40×. (J–L) Anterior is to the upper left. Magnification is 40×. Cells with strong myc staining are in a +/+ background; cells homozygous for usp mutants are myc negative and are marked with dotted lines. (H, K) The morphogenetic furrow is marked with an arrow.
The usp2 and usp3 phenotypes diverge in the region posterior to the furrow, a region in which BrC-Z1 is normally activated. In usp3 and usp4 clones, post-furrow activation of BrC-Z1 is maintained. Higher levels of BrC-Z1 occur posterior to the furrow relative to what is seen in the anterior part of a usp3 clone (Fig. 3 J–L, data not shown). Because loss of usp accelerates developmental events (19), such posterior BrC-Z1 expression occurs prematurely in mutant clones relative to wild-type tissue (Fig. 3 K and L). In usp2 clones, this additional level of BrC-Z1 activation is not evident posterior to the furrow (Fig. 3 G–I). These experiments show that USP is necessary for a component of BrC-Z1 activation and that USP3 retains this activation ability, consistent with the EcR results. If BrC-Z1 regulation normally occurs via EcR/USP heterodimers, our results suggest that receptor complex activation can mechanistically differ from repression in terms of DNA binding by the USP component.
Wild-Type USP Suppresses BrC-Z1 Expression in usp Clones.
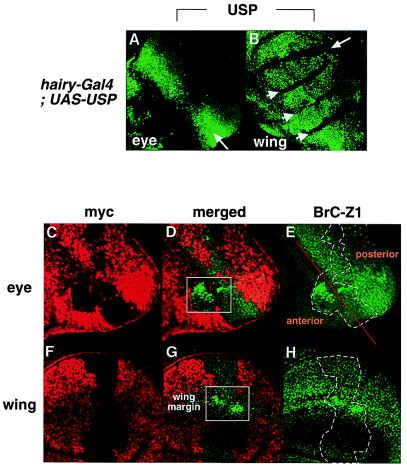
To verify that usp represses BrC-Z1, we used the Gal4-UAS system to ask whether targeted expression of USP could rescue BrC-Z1 repression (25). In the eye disc, we used hairyIJ3-Gal4 to drive USP expression to levels above wild type just anterior to the morphogenetic furrow. Under this driver, high levels of USP expression persist within and posterior to the furrow (Fig. 4A). In a usp mutant clone spanning the furrow (Fig. 4 C–E), expression of BrC-Z1 is high in a portion of the clone mostly anterior to the hairy domain of expression (Fig. 4D, boxed area) but is mostly rescued by exogenous USP in the furrow and post-furrow regions (Fig. 4 D and E).
Figure 4.
Position-specific ectopic USP expression restores BrC-Z1 expression to normal levels. An eye disc (A) and a wing disc (B) were isolated from hairyij3-Gal4/+; UAS-USP/+ larvae and stained with the anti-USP antibody (FITC). In the eye (A), ectopic USP, under control of the hairy-Gal4 driver, is expressed in a domain beginning anterior to the furrow (arrow) and continuing into the posterior of the disc. Anterior is to the bottom left. In the wing (B), exogenous USP is expressed throughout most of the disk except for a domain spanning the future wing margin (arrow). Arrowheads mark folds in the wing disc, which are not in focus but still contain ectopic USP. An eye disc (C–E) and a wing disc (F–H) containing usp3 clones are stained with anti-myc antibodies (Texas red) and with anti-BrC-Z1 antibodies (FITC). Clonal areas for usp3 are marked with dotted lines (E, H); an orange line marks the morphogenetic furrow (E). Regions where BrC-Z1 remains up-regulated are outlined with boxes (D, G). Magnification for all tissues is 40×.
In the wing imaginal disc, hairyIJ3-Gal4 drives USP expression throughout the disk, with the exception of the future wing margin region (Fig. 4B, arrow). In the usp3 clone shown (Fig. 4 F–H), where exogenous USP is expressed, BrC-Z1 expression is repressed to normal levels, except where the clone intersects the hairy nonexpressing region of the wing margin (Fig. 4G, boxed area).
DNA-Binding Properties of USP Alleles.
Although usp3, usp4, and usp2 have similar lethal phenotypes and fail to repress BrC-Z1 (Fig. 3), they differ in their ability to activate transcription of EcR and BrC-Z1 (Fig. 2). One possibility is that wild-type USP DNA-binding properties are required for repression but not for activation of at least some targets. As a first step in addressing this question, we examined the DNA-binding properties of wild-type and mutant USP proteins.
In our gel shift analysis, we found that wild-type USP binds various direct repeat sites as a monomer, as the USP/DNA complex is the same for these sites as for the half-site (Fig. 5A, lanes 3, 5–9). However, USP is able to bind a direct repeat with a spacer of 1 nucleotide (DR1) as a homodimer (Fig. 5A, lane 4), similar to what is seen with RXR (32). Unlike RXR, USP does not require ligand association to bind to the DR1 element. USP3 and USP4 mutant proteins were unable to bind DR1 or other elements (Fig. 5B, lanes 3 and 4). Because USP and its variants are translated equally well in vitro (Fig. 5C), the failure of mutant USP proteins to bind DR1 is likely because of their compromised DNA-binding properties. To confirm these DNA-binding properties, we tested the ability of VP16:USP fusion proteins (Fig. 1B) to activate transcription from an array of DR elements in cultured cells. Wild-type USP:VP16 activates the DR1-tk-luc reporter (Fig. 5D, column 2), but VP16:USP3 and VP16:USP4 fail to activate (Fig. 5D, columns 3 and 4), in agreement with the in vitro DNA-binding results. Activation of the DR1 element, on which USP can form homodimers, is independent of the ligand-binding domain (LBD) as a VP16:USP DBD, containing only the DBD region (amino acids 50–205) of USP, also activates DR1 (Fig. 5D, column 5).
USP Binding Sites Can Mediate Repression in Drosophila Cells.
As DR1 is a potential target element for USP, we next investigated whether DR1 mediates repression in Drosophila cells containing endogenous USP (12, 13). Transfection experiments were carried out by introducing reporters with different numbers of DR1 elements into Drosophila Schneider 2 cells. As a positive control for repression, we also tested the hsp27EcRE, an ecdysone-responsive element, which has been shown to mediate potent transcriptional repression in Drosophila cells (14, 15, 33). We found that DR1 sites could mediate repression and that repression increases with the number of DR1 elements in the reporter (Fig. 5E, columns 2 and 3). Repression mediated by three copies of DR1 is comparable to that of three copies of hsp27EcRE (Fig. 5E, column 4), strongly suggesting that the USP-binding DR1 elements can act as repressive sites in Drosophila cells.
USP3 and USP4 Activate Transcription in the Presence of EcR.
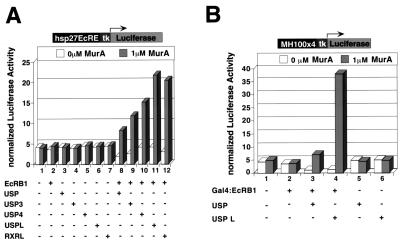
We investigated the basis of the USP3 and USP4 activation functions in cell culture. As a target element, we used the hsp27EcRE, which responds to ecdysone in both Drosophila and vertebrate cells. In these experiments, we used a potent EcR agonist, muristerone A, as a substitute for ecdysone (34). When CV-1 cells were cotransfected with the hsp27EcRE reporter and plasmids encoding the B1 isoform of EcR and wild-type USP, moderate activation of hsp27EcRE was observed (Fig. 6A, column 8). Activation was also observed when plasmids encoding USP3 and USP4 were used (Fig. 6A, columns 9 and 10). These results are consistent with our in vivo observation of activation of ecdysone response genes in usp3 and usp4 clones. To further determine whether DNA binding of USP is indeed dispensable for activation by EcRB1, we used a USP variant lacking the DBD but containing the LBD (USPL). In cells expressing USPL and EcRB1, activation still occurs and at levels higher than wild type (Fig. 6A, column 11). A similar result was obtained with the RXR LBD alone, suggesting that USP and RXR use a similar mechanism to contribute to activation by EcR (Fig. 6A, column 12). That activation by USPL and EcR results from a direct interaction between the two proteins is supported by the fact that they strongly interact in yeast two-hybrid assays (not shown). A related result consistent with the yeast data uses a heterologous Gal4 reporter, MH100x4 (Fig. 6B) in mammalian cells. In conjunction with Gal4-EcRB1, USPL is able to promote considerable activation of the reporter (Fig. 6B, column 4).
Figure 6.
USP3 or USP4, along with EcRB1, activates hsp27EcRE. Various wild-type and mutant USP derivatives were tested for their ability to confer, in conjunction with EcR, responsiveness to the EcR agonist muristerone A (34). (A) CV-1 cells were cotransfected with a reporter, hsp27EcRx5-tk-luc, and expression plasmids corresponding to EcRB1 (lanes 1, 8–12), USP (lanes 3, 8), USP3 (lanes 4, 9), USP4 (lanes 5, 10), USPL (lanes 6, 11), or RXRL (lanes 7, 12). hsp27EcREX5-tk-luc is an ecdysone response reporter containing five copies of hsp27EcRE. USPL or RXRL is a truncated protein encoding the LBD domain only. (B) CV-1 cells were cotransfected with plasmids expressing Gal4-EcRB1 (lanes 2–4), USP (lanes 3, 5), or USPL (lanes 4, 6). Gal4-EcRB1 is a Gal4 fusion protein, in which EcRB1 is fused C-terminal to the Gal4 DBD module. pMH100-tk-Luc is a Gal4 response reporter, containing four copies of the yeast Gal4 binding element.
Discussion
Our data suggest a separation between the repressive functions of USP and some of its activating functions as the USP DBD is dispensable for the activation of some ecdysone targets. usp3 and usp4 are capable of heterodimerizing with EcR, although they are defective in DNA binding (31). Here, we present evidence that on some EcREs, USP3 or 4/EcR heterodimers mediate activation. In culture, the USP LBD alone seems sufficient for the formation of an activating complex with EcR. Thus, for some genes, models explaining USP/EcR gene activation must accommodate the fact that the USP DNA-binding domain is not necessary, whereas the LBD is. One such model posits that EcR monomers, homodimers, or alternative EcR complexes can bind some EcREs but only activate if the USP LBD is present to promote formation of the EcR complex, ligand-binding, and/or interaction with coactivators. This model suggests that USP3, USP4, USPL, and possibly USP+ can activate through a multimeric complex in which the LBDs heterodimerize and DNA binding occurs largely via one or more EcR DBDs. In support of this model, ligand-induced EcR homodimers are able to form on DNA (C.-C. T., unpublished results).
In contrast to activation, repression of BrC-Z1 clearly requires functional USP DNA-binding abilities, whereas its post-furrow activation in the eye imaginal disk does not. The apparent differential requirement for DNA binding in repression and activation suggests that, in some situations, the switch between repression and activation regulated by the EcR/USP heterodimer may involve more than just changes in the LBD in response to ligand. It is also possible that normally the switch from repression to activation occurs without a change in the DNA binding of either EcR or USP but that on some target sites in the absence of the wild-type complex, an alternative complex can form and allow activation.
The ability of added wild-type USP to restore BrC-Z1 repression in the eye imaginal disk suggests that the Z1 isoform of BrC may be a direct target of USP regulation. As we have shown that USP has the ability to homodimerize on a DNA element able to mediate repression in Drosophila cells, it is possible that an alternative USP complex other than EcR/USP represses BrC-Z1. If USP is able to repress target genes through a homodimer but requires heterodimerization with EcR to mediate activation, a situation could arise in which gene repression absolutely requires the DNA-binding activity of USP while this function can be abolished for gene activation.
In this study we have uncovered a dual role for USP in the ecdysone response. Depending on the particular target gene, activation and repression may be more complicated than just a simple ligand-activated switch. This adds potential complexity to the roles that ecdysone, USP, and EcR play in metamorphosis. Our work separates aspects of the USP component of the ecdysone response into repressive and activating functions, with unique and separable effects attributable to the DNA-binding and ligand-binding domains. Given conservation of the properties of EcR/USP heterodimers with vertebrate RXR-containing complexes, these results may be relevant to understanding aspects of regulation by RXR heterodimers in vertebrates.
Acknowledgments
We thank Dr. Greg Guild, Dr. F. Kafatos, and the Developmental Studies Hybridoma Bank for antibodies, and we thank the Bloomington Drosophila Stock Center for fly stocks. We are grateful to Dr. F. Gage for use of the confocal microscope. N.G. was supported by a predoctoral National Institutes of Health training grant. C.-C.T. is a fellow of the Damon Runyon-Walter Winchell Cancer Fund (DRG 1366). M.M. and R.M.E. are members of the National Institutes of Health cancer center at the Salk Institute. R.M.E. is an investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by grants from American Cancer Society and the U.S. National Science Foundation (to M.M.) and by the Howard Hughes Medical Institute, National Institutes of Health Grants HD27183 and GM26444, the Mathers Foundation, and the Joe and Dorothy Brown Dorsett Foundation.
Abbreviations
- USP
ultraspiracle
- EcR
ecdysone receptor
- RXR
retinoid X receptor
- BrC-Z1
the Z1 isoform of broad complex
- DBD
DNA-binding domain
- LBD
ligand-binding domain
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 2.Kliewer S A, Umesono K, Heyman R, Mangelsdorf D J, Dyck J A, Evans R M. Proc Natl Acad Sci USA. 1992;89:1448–1452. doi: 10.1073/pnas.89.4.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kliewer S A, Umesono K, Mangelsdorf D J, Evans R M. Nature (London) 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stunnenberg H G. BioEssays. 1993;15:309–315. doi: 10.1002/bies.950150504. [DOI] [PubMed] [Google Scholar]
- 5.Glass C K. Endocr Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 6.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 7.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 8.Xu L, Glass C K, Rosenfeld M G. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 9.Lemon B D, Freedman L P. Curr Opin Genet Dev. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 10.Oro A E, McKeown M, Evans R M. Nature (London) 1990;347:298–301. doi: 10.1038/347298a0. [DOI] [PubMed] [Google Scholar]
- 11.Koelle M R, Talbot W S, Segraves W A, Bender M T, Cherbas P, Hogness D S. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- 12.Yao T P, Segraves W A, Oro A E, McKeown M, Evans R M. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 13.Yao T P, Forman B M, Jiang Z, Cherbas L, Chen J D, McKeown M, Cherbas P, Evans R M. Nature (London) 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 14.Cherbas L, Lee K, Cherbas P. Genes Dev. 1991;5:120–131. doi: 10.1101/gad.5.1.120. [DOI] [PubMed] [Google Scholar]
- 15.Dobens L, Rudolph K, Berger E M. Mol Cell Biol. 1991;11:1846–1853. doi: 10.1128/mcb.11.4.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai C C, Kao H Y, Yao T P, McKeown M, Evans R M. Mol Cell. 1999;4:175–186. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- 17.Thummel C S. Trends Genet. 1996;12:306–310. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- 18.Karim F D, Thummel C S. EMBO J. 1992;11:4083–4093. doi: 10.1002/j.1460-2075.1992.tb05501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelhof A C, Ghbeish N, Tsai C, Evans R M, McKeown M. Development. 1997;124:2499–2506. doi: 10.1242/dev.124.13.2499. [DOI] [PubMed] [Google Scholar]
- 20.Schubiger M, Truman J W. Development. 2000;127:1151–1159. doi: 10.1242/dev.127.6.1151. [DOI] [PubMed] [Google Scholar]
- 21.Emery I F, Bedian V, Guild G M. Development. 1994;120:3275–3287. doi: 10.1242/dev.120.11.3275. [DOI] [PubMed] [Google Scholar]
- 22.Bayer C A, von Kalm L, Fristrom J W. Dev Biol. 1997;187:267–282. doi: 10.1006/dbio.1997.8620. [DOI] [PubMed] [Google Scholar]
- 23.Xu T, Rubin G M. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 24.Oro A E, McKeown M, Evans R M. Development. 1992;115:449–462. doi: 10.1242/dev.115.2.449. [DOI] [PubMed] [Google Scholar]
- 25.Brand A H, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 26.Jindra M, Malone F, Hiruma K, Riddiford L M. Dev Biol. 1996;180:258–272. doi: 10.1006/dbio.1996.0299. [DOI] [PubMed] [Google Scholar]
- 27.Talbot W S, Swyryd E A, Hogness D S. Cell. 1993;73:1323–1337. doi: 10.1016/0092-8674(93)90359-x. [DOI] [PubMed] [Google Scholar]
- 28.Christianson A M, King D L, Hatzivassiliou E, Casas J E, Hallenbeck P L, Nikodem V M, Mitsialis S A, Kafatos F C. Proc Natl Acad Sci USA. 1992;89:11503–11507. doi: 10.1073/pnas.89.23.11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umesono K, Evans R M. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 30.Zelhof A C, Yao T P, Chen J D, Evans R M, McKeown M. Mol Cell Biol. 1995;15:6736–6745. doi: 10.1128/mcb.15.12.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrich V C, Szekely A A, Kim S J, Brown N E, Antoniewski C, Hayden M A, Lepesant J A, Gilbert L I. Dev Biol. 1994;165:38–52. doi: 10.1006/dbio.1994.1232. [DOI] [PubMed] [Google Scholar]
- 32.Lee M S, Kliewer S A, Provencal J, Wright P E, Evans R M. Science. 1993;260:1117–1121. doi: 10.1126/science.8388124. [DOI] [PubMed] [Google Scholar]
- 33.Luo Y, Amin J, Voellmy R. Mol Cell Biol. 1991;11:3660–3675. doi: 10.1128/mcb.11.7.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christopherson K S, Mark M R, Bajaj V, Godowski P J. Proc Natl Acad Sci USA. 1992;89:6314–6318. doi: 10.1073/pnas.89.14.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]