Summary
Defects in closure of embryonic tissues such as the neural tube, body wall, face and eye lead to severe birth defects. Cell adhesion is hypothesized to contribute to closure of the neural tube and body wall, however potential molecular regulators of this process have not been identified. Here we identify an ENU-induced mutation in mice that reveals a molecular pathway of embryonic closure. Line2F homozygous mutant embryos fail to close the neural tube, body wall, face, and optic fissure, and they also display defects in lung and heart development. Using a new technology of genomic sequence capture and high-throughput sequencing of a 2.5 Mb region of the mouse genome, we discovered a mutation in the grainyhead-like 2 gene (Grhl2). Microarray analysis revealed Grhl2 affects the expression of a battery of genes involved in cell adhesion and E-cadherin protein is drastically reduced in tissues that require Grhl2 function. The tissue closure defects in Grhl2 mutants are similar to that of AP-2α null mutants and AP-2α has been shown to bind to the promoter of E-cadherin. Therefore, we tested for a possible interaction between these genes. However, we find that Grhl2 and AP-2α do not regulate each other’s expression, E-cadherin expression is normal in AP-2α mutants during neural tube closure, and Grhl2;AP-2α trans-heterozygous embryos are morphologically normal. Taken together, our studies point to a complex regulation of neural tube fusion and highlight the importance of comparisons between these two models to understand more fully the molecular pathways of embryonic tissue closure.
Keywords: embryonic morphogenesis, neural tube defects, neural tube development, tissue closure, E-cadherin, Grhl2, AP2α
Introduction
Neurulation, the process that gives rise to the central nervous system, in mammals is a complex and highly dynamic morphogenetic process during which the flat neural plate rolls up and closes to form the neural tube. Neurulation occurs in at least four distinct steps, namely thickening of the ectoderm to form a flat neural plate, elevation of the lateral edges of the neural plate to form the neural folds, apposition of the neural folds, and finally, joining of the neural folds in the midline. This last step includes separation of the neural ectoderm from the non-neural ectoderm and closing of these tissues to form the neural tube covered by a single layer of ectoderm. This final closure event has traditionally been called fusion, although the cells do not actually fuse. Neural tube defects (NTD) or failure of neural tube closure occurs in ~1:1000 live human births. Studies in mice have lent considerable insight into the genetics of neural tube closure and have identified genes involved in processes including neural patterning, cell proliferation, and tissue architecture, however, relatively little is known about the molecular basis of fusion of the neural folds (Harris and Juriloff, 2010). Other embryonic tissues also undergo closure events and it has been proposed that similar mechanisms of fusion are utilized. Body wall closure occurs when the lateral edges of the embryonic ectoderm and mesoderm are joined and fuse at the ventral midline. Defective body wall closure can lead to an open abdominal and/or thoracic cavity. Closure of the optic fissure occurs through joining of the folds of the optic cup, disruption of which results in coloboma. Finally, the facial prominences also undergo fusion and defects in facial closure can result in birth defects ranging from cleft lip and palate to clefting of the entire face. Although birth defects due to failure of closure of embryonic tissues are of considerable medical consequence, the cellular and molecular events that regulate fusion remain unknown.
It has been long hypothesized that fusion relies on adhesion molecules for its completion. Although this is a compelling idea, the molecules that regulate the process of neural tube, body wall or optic fissure closure are not well defined. With respect to neural tube closure, mutations in adhesion molecules, such as EphrinA5 and EphrinA7, can cause NTD in mice (Holmberg et al., 2000) although neither has been shown to be directly involved in the step of neural fold fusion. Cadherins are also thought to be predominant cell adhesion molecules involved in initial recognition and formation of cell-cell interactions between the juxtaposed neural folds during mammalian neural tube formation. Two observations that support this hypothesis are different subtypes of cadherins control various aspects of embryonic development (Takeichi, 1988), and subtypes of cadherins are essential for neurulation in a number of animal models, including Xenopus (Rashid et al., 2006) and mice (Chen and Hales, 1995). However, the details of their function and what step of neurulation these molecules control are not fully understood. No specific adhesion molecules are yet implicated in closure of the body wall. The AP-2α null mouse is one example of a gene mutation that causes defects in body wall and neural tube closure (Brewer and Williams, 2004; Nottoli et al., 1998). In cancer cell lines, AP-2α has been shown to bind the promoter of E-cadherin and the functional consequences of this have been studied in adult tissues and cancer cells (Baldi et al., 2001; Schwartz et al., 2007). The relationship between AP-2α and E-cadherin in the embryo has only been explored in the cornea of the developing eye (West-Mays et al., 2003).
Here we describe the identification of an ENU-induced nonsense mutation in the grainyhead-like 2 (Grhl2) gene that encodes a transcription factor. This mutation reveals the requirement for Grhl2 in closure of embryonic tissues as Grhl2 mutants display cranial NTD (exencephaly), cleft face, body wall closure defect (thoracoabdominoschisis), and defective optic fissure closure (coloboma). Moreover, Grhl2 is required in other tissues as mutant embryos display lung and heart defects, soft tissue syndactyly, and molecular changes in the skin and hair follicle. Microarray analysis comparing wildtype and mutant cranial tissue reveals changes in a number of genes that encode proteins involved in cell adhesion. Immunohistochemical analysis demonstrates a dramatic reduction in E-cadherin expression in mutant tissues, corresponding to where Grhl2 would normally be expressed. Moreover, we explore the relationship between the Grhl2 and AP-2α transcription factors during neural tube closure. Despite the similar fusion defects, Grhl2 and AP-2α do not regulate one another’s transcription and AP-2α is not required for E-cadherin protein expression. Thus, comparisons between these two models will be important in uncovering the molecular complexity that underlies the regulation of key embryonic closure events.
Materials and methods
Generation of Line2F embryos and genetic mapping
The Line2F mutation was induced with the chemical mutagen ethyl nitrosourea (ENU) on a C57BL/6J genetic background. Originally, the line was outcrossed to C3H3B/FeJ and later outcrossed to 129S1/SvImJ (129) for 7 generations. Meiotic mapping placed the Line2F mutation on mouse chromosome 15 between initial markers D15Mit8 (33,393,085 bp) and D15mit138 (39,861,164 bp) and was further defined by D15Am33.45A (forward primer CGT CTA GAG GCA CTG TGA GG, reverse primer TTT CCC TGG AAA CAC TAA GTCAA). This marker lies at 36,643,830 bp and the number of recombination events over the number of opportunities for recombination was 2/50. Markers that were also used to further define the region were: D15MIT49 (37,204,637 bp; 0/170), D15MIT6 (38,474,396; 17/207), and FA6 (39,430,000; 25/207) forward primer TCC AAA CTG TTT TAA AGG CAA A, reverse primer CAA CTT GAA TAT TGT ATG CAT CTC C).
Heterozygous carriers were mated and the embryonic stage was determined considering noon of the day of a vaginal plug as E0.5. At dissection, the embryonic phenotype was recorded and a small portion of the embryo or yolk sac removed for genotyping. Embryos were fixed in 4% paraformaldehyde (PFA) overnight and then processed for whole mount or section (10μm) in situ hybridization or for immunohistochemistry.
Sequence capture and 454 sequencing
A targeted sequence capture strategy was used to preferentially enrich for and subsequently sequence the critical genomic region containing the Line2F mutation to identify the genetic basis of the mutant phenotype. Roche-Nimblegen custom designed hybrid capture microarray chip with 385,000 spotted oligonucleotides was used to capture and enrich for the region of interest. Based on the C57BL/6 mouse reference genome sequence, oligonucleotides were designed by Roche-Nimblegen that covered the 36.0Mb to 38.5Mb target region on mouse chromosome 15 but avoided repetitive element sequences. After the initial design phase, we slightly lowered the stringency of exclusion of repeat elements so that coverage of more genic and intergenic regions would be provided by the tiling scheme.
For DNA preparation, E10.5 embryos from a heterozygous cross were dissected and the yolk sac used to determine the genotype. Three homozygous mutant embryos from the same litter were homogenized with a handheld mechanical homogenizer. DNA extraction was performed using Qiagen DNA extraction kit (cat#69581), which yielded ~10 μg of DNA from three E10.5 embryos. These embryos were homozygous for any potential mutations within this 2.5Mb region and this region would derive from C57BL/6. Nimblegen processed the genomic DNA using fragmentation by nebulization, size selection, end-repair, and linker addition to the ends. This genomic library was then hybridized to the custom-designed hybridization microarray chip using the standard Nimblegen sequence capture protocol. In brief, this protocol (conducted by Nimblegen) involved hybridization to the chip, washing away non-hybridized sequences, elution of hybridized library from the chip, and evaluation of enrichment via qPCR. The resulting enriched library was further processed to be compatible with 454 sequencing by ligation of 454-specific adapters and selection with beads for the correct compliment of adapters. The enriched library was sequenced by the Consortium for Comparative Genomics at the University of Colorado Denver using FLX emulsion PCR kit 1. Enriched emulsion PCR beads were sequenced on the 454 GS-FLX sequencer using 70×75cm sequencing plates and FLX-LR sequencing reagents. All emulsion PCR, bead enrichment, and sequencing methods strictly adhered to default 454-provided protocols.
Analysis of Sequencing Results
The data were analyzed using the 454 GS-mapper software by the Consortium for Comparative Genomics at the University of Colorado Denver with assistance by V. Vemulapali. The targeted region of the mouse genome from C57BL/6 strain was downloaded from the UCSC browser and used as a reference for mapping of 454 sequence reads. Default parameters were used for 454 GS-mapper software analyses. Approximately 80% of the sequence obtained from the enriched library mapped back to the region of interest. The GS-mapper produced a file with the details of high quality mutations, and this file was further processed and summarized using a Perl script. Because our source genomic DNA was expected to be homozygous for any mutations within the targeted region, the Perl script filtered out putative mutations at sites where at least one read matched the reference sequence. Sequencing data from the 454 GS-FLX sequencer were aligned with the published C57BL/6 sequence using the 454 sequencer software to generate a list with nucleotides that differed in the mutant DNA (referred to as SNPs). Further analysis of the SNPs was done through Ensembl (http://uswest.ensembl.org/Mus_musculus/Info/Index) and the Expasy translation engine (http://www.expasy.ch/tools/dna.html) to determine the effect of the nucleotide change on predicted proteins and potential regulatory elements.
These analyses identified a SNP in the coding region of Grhl2 predicted to create a stop codon. To verify this result, DNA from 4 mutant and 4 wildtype embryos was extracted using Qiagen DNA extraction kit (cat#69581). A 400-nucleotide region around position 37,200,529 bp on chromosome 15 was PCR amplified with custom designed primers (Sequencing Forward: AAA GAC TAG TGG CCT TAG TGC CCA; Sequencing Reverse: CTC GGT GAT GGA TAC ACT GTA CTG CT). Sequencing of the 400 nucleotide region was performed at the Barbara Davis Diabetes Center Molecular Biology Core using these same custom primers.
For genotyping of carrier animals and embryos, the Grhl21Nisw mutation was specifically detected using ARMS primers according to Ye et al. (2001): Outer primer 1: TAG TGG CCT TAG TGC CCA TGC CCA GTG; outer primer 2: TGA AGT TGG GTT TCA CAA GGG GGG AGG; A allele primer: CTG CCG CCC TGG GCC TGC TCA AA; T allele primer: CCA AGG GAG AGC CCA CCT TGT AGT AGA CA.
Microarray and data analysis
E9.5 mouse embryos (~23–25 somites) were collected and tissue from the head above the first branchial arch was dissected, frozen in dry ice and stored at −80°C, while the remainder of the embryo was used for genotyping to distinguish mutant from wildtype. Tissue from eight homozygous mutant and eight homozygous wild type embryos from four different litters was collected and then processed at the same time for consistency purposes. RNA was extracted using RNA-easy micro kit (Qiagen) and kept at −80°C until use.
RNA amplification, labeling and hybridization for the microarray experiment was performed according to the Affymetrix protocol as described in the user manual GeneChip®Whole Transcript (WT) Sense Target Labeling Assay, and the microarray chip used for the hybridization was from Affymetrix Mouse Gene 1.0 ST version 1. The microarray experiments were conducted by the University of Colorado Denver Microarray Core Facility.
Analysis of the microarray data was performed in collaboration with T. Shade from the UC Denver microarray core. All available raw gene expression data (probe-level) from Affymetrix CEL files was imported into Partek Genomics Suite (version 6.4, build 6.08.1010, copyright 1993–2008, Partek Inc., St. Louis, MO, USA) software for data analysis, i.e. Pre-background Adjustment, Background Correction, Normalization, and Summarization. Principal Component Analysis PCA was performed on the normalized data to identify any outlier samples. Prior to performing statistical analysis, the control probes were filtered out, leaving only the interrogating probes. No other filtering was performed on the dataset prior to performing the Analysis of Variance ANOVA to determine statistically significant gene regulation between the two experimental groups. The data was log base 2 (log2) transformed prior to running the ANOVA. After the ANOVA, the log2 ratios were converted into a linear scale fold change. P-values were calculated determining the most statistically significant gene changes. The dataset was then limited to genes with fold changes higher than 1.25 to observe the most robust up- and down-regulated genes for the two experimental groups.
Tissue for quantitative real-time PCR was collected and RNA was prepared as for the miccroarray experiment (see above). cDNA was made with the use of SuperScript III first strand kit (Invitrogen cat# 18080-051) and qRT-PCR was performed with TaqMan probe-based gene expression analysis, using primers commercially available from Applied Bioscience.
RNA in situ hybridization and immunofluorescence
Whole-mount and section RNA in situs were performed as described (Holmes and Niswander, 2001; Liu et al., 1998). Immunohistochemistry on cryosections was performed as described (Kim et al., 2007). Antibodies: Islet1 (Developmental Studies Hybridoma Bank 39.4D5; dilution 1:100), TuJ1 (Convance cat# MMS-435P; dilution 1:1000), Phospho-histone3 (Cell Signaling cat#9701-S; dilution 1:200), E-cadherin and Ezrin (Transduction laboratories; both at 1:100), Beta-catenin (Santa Cruz biotechnology; 1:50). Immunofluorescence was visualized with LSM510 META confocal microscope from Zeiss.
Proliferation
Transverse cryosections at the level of the rostral spinal cord/caudal hindbrain from E8.5, E9.5 and E10.5 mutant and wild type embryos were stained with antibodies against phospho-Histone3 (1:200 dilution), phalloidin (1:400), Hoechst (1:1000). Five embryos from each age and ~6 sections/embryo were used for cell counting. The number of dividing cells in the neural tube of each section, as determined by the presence of phospho-Histone3 staining, was divided by the number of total cells in the neural tube per section as determined by the Hoechst nuclear stain to quantify the percentage of proliferating cells/total cells in each section. The average of this ratio was compared using unpaired two-tailed student’s t-test between mutant and wild type. For all three ages, such a comparison lead to non-significant p value > 0.05.
Results
Genomic sequence capture and high throughput sequencing identify Grhl2 mutation in Line2F embryos with defective tissue closure
Using ENU mutagenesis and a three-generation forward genetic screen as an unbiased approach to identify recessive genes that are critically required for neural tube closure, we identified Line2F. As described in the next section, Line2F homozygous mutant embryos displayed abnormalities in many tissues including failure to close the neural tube, face, body wall, and optic fissure, as well as soft tissue syndactyly, lung and heart defects. To identify the ENU-induced mutation, we used meiotic recombination mapping to determine association between the mutant phenotype and the C57BL/6J genetic background on which the mutation was generated. This defined the Line2F mutation to a less than 2 Mb region on chromosome 15 between 36.64 and 38.51 Mb. Within this interval are 11 transcription units, as listed in Supplemental Table 1, of which one is a predicted gene and the null phenotype of four other genes are not compatible with the Line2F phenotype as they die later than Line2F mutants and they do not have NTD (An et al., 2010; Kimura et al., 2003; Wan et al., 2010; Zhou et al., 2007)
The traditional approach to identify the causative mutation induced by ENU mutagenesis has been high-resolution mapping, followed by amplification and sequencing of coding regions and splice junctions of genes in the interval. Although the current positional cloning process is relatively straightforward, it does require expertise in mouse genetics, ample vivarium space and considerable genotyping, which translate to high cost. Moreover, the process is labor intensive since it relies on PCR amplification of large numbers of individual templates that are then sequenced, further adding to the cost, in particular if the critical region is gene-rich and/or has no obvious candidate genes. Furthermore, traditional sequencing for mutations in genes in the interval or exome sequencing focus on sequencing of coding sequences and splice junctions and hence may be fruitless if the mutation lies in noncoding sequence or sequence that is not well annotated.
Here we used a new technology of genomic sequence capture followed by high-throughput sequencing to identify in an unbiased way the ENU-induced mutation(s) throughout this interval, including in regulatory elements and sequences between genes or exons. Using this approach, all potential mutations within a candidate region as large as 5Mb can be found in a few months. Thus, sequence capture technology can significantly reduce the time and resources required for mutation identification because it eliminates the need for high resolution genetic mapping, long-range PCR, and sequencing of individual PCR amplimers. The sequence capture approach has been previously used to amplify a 200 kilobase region in order to identify novel alleles in mutant models of the Kit locus (D’Ascenzo et al., 2009). In our study, we used this technique for the first time to identify an ENU-generated mutation in a much larger area of the genome. We captured a region that was ~12.5 times greater to identify the causative ENU-induced mutation in Line2F.
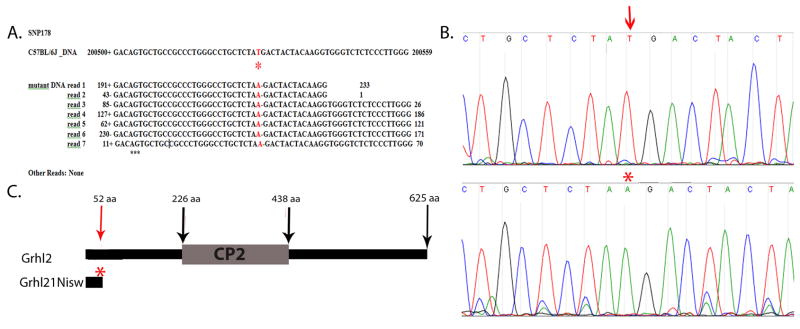
As described in more detail in the Methods section, closely spaced probes that targeted unique sequences between 36.0–38.5Mb on mouse chromosome 15 were designed by Roche Nimblegen and placed on a microchip. Genomic DNA was extracted from homozygous mutant embryos, fragmented, and hybridized to the custom primer array to enrich for this 2.5Mb region. Approximately 80% of the recovered DNA mapped back to the region of interest, demonstrating successful enrichment. The enriched DNA was sequenced using 454 GS-FLX sequencing to ~20X coverage. Comparative analysis of these sequences against the mouse C57BL/6 reference genome showed only one single mutation, SNP178, that altered the amino acid translation of a coding region. SNP178 was consistent in all seven reads from the mutant embryo and there were no other reads (Fig. 1A).
Fig. 1. Sequence capture identifies Grhl2 mutation in Line2F.
Sequence capture to enrich for genomic sequences from 36.0–38.5 Mb on mouse chromosome 15 followed by high-throughput sequencing identified (A) SNP178 in which all reads from homozygous mutant DNA show a consistent change from T to A (red asterisk) that differs from the publically available C57BL/6 sequence. This is predicted to lead to an early stop codon in the Grhl2 gene.
(B) Verification of the base change was done by PCR amplification of 400bp of Grhl2 from DNA from wildtype (top) and mutant (bottom) embryos followed by Sanger sequencing. The chromatogram shows the T (red arrow) to A (red asterisk) nucleotide change.
(C) Schematic of Grhl2 protein. The Grhl21Nisw mutation is predicted to truncate the protein 52 amino acids from the N-terminus of the protein and to delete the CP2 DNA binding domain.
SNP178 represented a T to A transversion at position 37,200,529 on chromosome 15 within the coding region of the gene grainyhead-like 2, Grhl2. This mutation was verified by using locus-specific primers to PCR amplify a 400bp fragment from four wildtype and four mutant embryos followed by Sanger sequencing using the same custom primers (Fig. 1B). This change in the mutant is predicted to introduce a premature stop codon at amino acid 52, and hence delete 573 amino acids from the predicted 625 amino acid protein normally encoded by Grhl2, including the predicted CP2 DNA binding domain (Fig. 1C) (Tuckfield et al., 2002). This allele will be called Grhl21Nisw.
The founding member of the Grhl family of transcription factors is Drosophila grainyhead (also called NTF-1 or Elf-1). The five mammalian Grhl family members form two distinct phylogenetic groups according to sequence alignment (Wilanowski et al., 2002). The first group consists of CP2, LBP-1a and LBP-9 genes, which are widely expressed and have diverse functions in T-cell proliferation, globin gene expression and steroid biosynthesis (Volker et al., 1997; Zhou et al., 2000). The second group consists of Mammalian Grainyhead or Grhl1, Brother-of-MGR or Grhl2, and Sister-of-MGR or Grhl3 (Ting et al., 2003b; Wilanowski et al., 2002). Grhl1 null mutations have defective hair growth (Wilanowski et al., 2002). Grhl3 null mutants display 100% penetrant spina bifida and ~3% exencephaly (Ting et al., 2003b). Grhl2 mutations are associated with hearing loss in humans (Van Laer et al., 2008). There have been two recent reports of mutations in Grhl2 in mice leading to exencephaly and split face (targeted allele, Rifat et al., 2010; gene trap alleles, Werth et al., 2010), similar to the phenotype discussed below for Line2F mutants. Although we have not provided definitive proof that the mutation in Grhl2 is causative for the Line2F phenotype, such as non-complementation in a genetic cross with another Grhl2 allele, we present strong evidence that suggests that it is a mutation in Grhl2 that causes the Line2F phenotype. This includes meiotic mapping of the critical interval in Line2F to less than 2Mb, and sequencing of this entire region, plus another 0.5Mb of flanking DNA, which showed the only mutation in a coding region was in the Grhl2 gene and this is predicted to lead to severe truncation of the Grhl2 protein. Moreover, presence of NTD and loss of E-cadherin expression (see below) in both the gene-trap Grhl2 mutants and the ENU-induced Grhl2 mutant, lend strong support to the idea that the Grhl2 mutation is responsible for the phenotype in Line2F.
Grhl21Nisw embryos show a general defect in closure of embryonic tissues
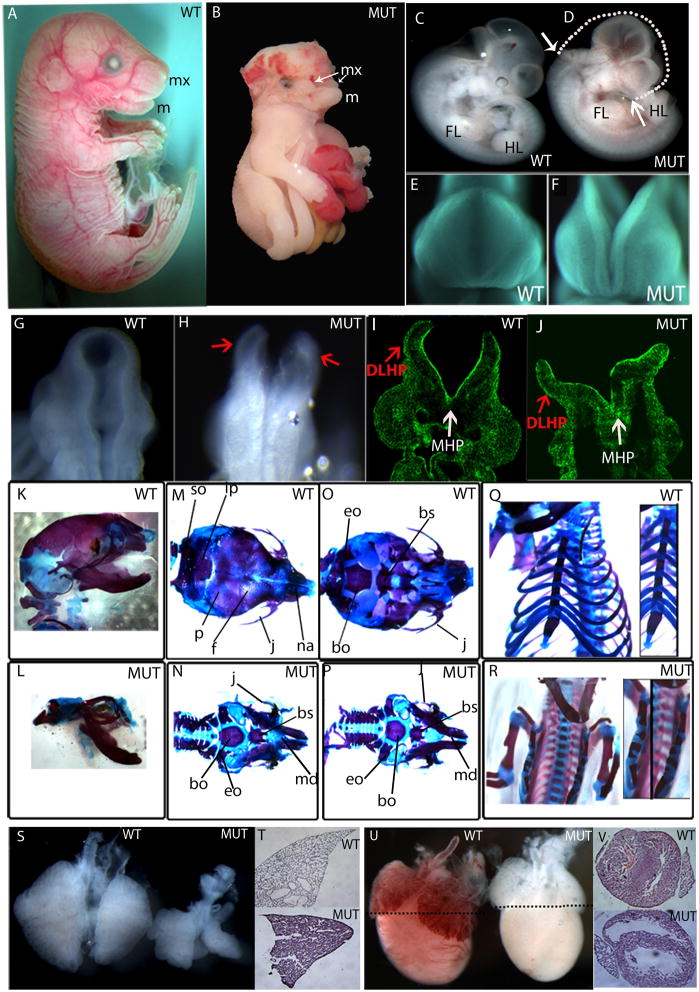
The ENU-induced Grhl21Nisw mutation was produced on a C57BL/6J genetic background. All of the analyses here were conducted on animals bred into 129S1/SvImJ (129) background for seven generations, except for the skeletal stainings, which were done on C3HeB/FeJ (C3H) background. On a 129 background, Grhl21Nisw/1Nisw embryos display complete exencephaly (hindbrain, midbrain, forebrain) and cleft face (100%) wherein the upper jaw (maxilla, mx) and upper part of the face fail to close (Fig. 2B, D, F compared to wild type in Fig. 2A, C, E), but the lower jaw (mandible, m) is formed (Fig. 2B; Supplemental Fig. 1A, B). Primary thoracoabdominoschisis is also observed (100% of the embryos that survive to E12.5 or later) exposing the organs within the thorax and abdomen to the uterine environment due to failure of body wall closure (Fig. 2B, Supplemental Fig. 1B). There is a high percentage of embryonic lethality at embryonic day E9.5 and only a few embryos (~5%) survive past that age. A small percentage (~5%) of mutant embryos displayed a more severe phenotype being significantly smaller than littermates, developmentally delayed and unturned, with open “wavy” neural folds (Supplemental Fig. 1C).
Fig. 2. Grhl2Nisw1 embryos show a general defect in closure of embryonic tissues and lung and heart defects.
(A–J) Wildtype (WT) and homozygous Grhl21Nisw mutant (MUT) embryos show a lack of closure of the neural tube and craniofacial region. Side view of E18.5 embryos (A, B) wildtype and mutant embryos showing the mutant is smaller with hind- to forebrain exencephaly, thoracoabdominoschisis, and soft tissue syndactyly. The mandible (m, lower jaw) is formed in both wild type and mutant embryos, however the maxilla (mx, upper jaw) fails to form properly in the mutant embryos and remains as two separate processes. Side view of E11.5 (C, D) embryos showing similar body size but NTD from hindbrain to forebrain and cleft upper face. White arrows on the mutant show the extent of the neural tube opening, while the white dotted line on the mutant indicates what would be the ~normal shape of the embryo head. Front (E and F) and back (G, H) view of the cranial region of E9.5 embryos showing failure of closure of the facial region and neural tube. (I, J) Section from E9.5 mutant embryo stained with phalloidin, which binds F-actin. Red arrows point to dorsolateral hinge points (DLHP), and white arrows point to medial hinge point (MHP), both of which form in the mutant.
(K–R) Skeletal stainings of E18.5 wildtype and mutant embryos on C3H background. Side (K), top (M), bottom (O) view of the skull and frontal view of the rib cage (Q) of wildtype embryos.
Side (L), top (N), bottom (P) view of the skull and frontal view of the rib cage (R) of mutant embryos.
(S) Wildtype and mutant lungs of E14.5 embryo. (T) H&E staining of E18.5 lung of wildtype and mutant embryos. (U) E18.5 heart of wildtype and mutant embryo. Black dotted lines indicate the level of section shown in (V).
so: supraoccipital, p: parietal, ip: interparietal, f: frontal, j: jugal, na: nasal, eo: exoccipital, bo: basioccipital, bs: basisphenoid.
Further outcrossing into 129 background increased the percentage of non-maintained pregnancies. Dissections of pregnant females later than 10.5 days post coitum (>5 generations outcross to 129) yielded small litters of approximately 2–3 embryos and a high number of resorptions (~3–6 resorbed embryos per litter). Based on Mendelian inheritance, this is a greater number than expected for loss of homozygous mutants, and hence suggests an additional defect, perhaps loss of heterozygous embryos or overall fetal loss perhaps due to a maternal defect.
On the C3H background, Grhl21Nisw/1Nisw embryos displayed exencephaly (100%), body wall defects (50%), cleft face or unilateral or bilateral cleft (100% in total) and soft tissue syndactyly (webbing between the digits, ~7%) and a proportion of these embryos survived to E18.5. Skeletal stainings of E18.5 mutant embryos showed lack of skeletal components of the top of the skull and midface (compare Figs. 2L, N to K, M) whereas the lower jaw and ventral skull were present (Fig. 2O, P). In the mutant thorax, the ribs attached normally to the sternum but the left and right sides of the sternum failed to connect in the midline to form one continuous bone (Fig. 2R, Q). Grhl21Nisw mutants also displayed eye defects that resemble the human condition called coloboma, wherein the edges of the optic fissure fail to close, leaving a gap in the iris (Supplemental Fig. 1D, E). Taken together, the lack of closure of the neural folds, craniofacial structures, body wall and optic fissure suggest that Grhl21Nisw mutants have a defect in the process of embryonic tissue fusion.
The neural tube closure defect in Grhl21Nisw is evident as early as E9.5 in that the neural folds had elevated normally and the folds were juxtaposed but not fused (Fig. 2G, H). This was not due to developmental delay by comparing somite-matched littermates and as dissection at later stages showed no evidence of closure. Bending of the neural folds appeared normal and histological sections show formation of the medial and dorsolateral hinge points in the mutant (Fig. 2I, J). Similarly, there is normal craniofacial growth and juxtaposition of the facial prominences at the midline but failure to close (Fig. 2E, F). These results differ from a recent report that described a targeted deletion in Grhl2 in which these embryos displayed a broadly open cranial neural tube that lacked dorsolateral hinge points and had minimal bending at the medial hinge point (Rifat et al., 2010). Additional phenotypic differences between the targeted mutation and the ENU Grhl21Nisw allele (on C57BL/6 and129 backgrounds, respectively) exist with respect to closure of the caudal neural tube. The posterior neuropore (PNP) was broadly open in the targeted allele and this remained open in all homozygotes (Rifat et al., 2010) whereas, in Grhl21Nisw mutants, the neural folds of the PNP were elevated (Supp. Fig. 1F, G) and we never detected a failure of PNP closure in older Grhl21Nisw embryos. Moreover, the body wall is apparently closed in the targeted allele whereas the body wall and optic fissure are not closed in the ENU allele. Between the different alleles, molecular differences are not expected. The Grhl21Nisw allele is expected to severely truncate the protein, including deleting the predicted CP2 DNA binding domain. Grhl2 RNA expression in Grhl21Nisw mutant embryos when examined by in situ hybridization appears to be localized properly and at levels comparable to wild type. Using a more quantitative method, qRT-PCR, however there is a 2.1 fold reduction in Grhl2 mRNA in the mutant (Table 1). The allele described by Rifat et al. (2010; C57BL/6 background) is a targeted mutation of Grhl2 and is expected to be a null allele, whereas the alleles studied in Werth et al. (2010; mixed 129P2/C57BL/6 or 129/ola/C57BL/6 background) derive from gene-traps of Grhl2 and are indicated to disrupt Grhl2 RNA and protein expression. Therefore, it is possible that the phenotypic differences arise from differences in genetic background. As we described above, the genetic background affects the penetrance of the defects and age of embryonic lethality in Grhl21Nisw mutants. At this time it is unclear whether the phenotypic and neural hinge point differences reflect the differences in the Grhl2 mutations and/or the genetic background on which the phenotypes were examined.
Table 1.
Gene expression tested by qRT-PCR
| Gene Symbol | Fold-Change (mutant vs. WT) | p-value (mutant vs. WT) | |
|---|---|---|---|
| 1 | Rab25 | 23.28 | 0.0231 |
| 2 | EpCam | 6.09 | 0.0463 |
| 3 | Esrp1 | 4.33 | 0.0151 |
| 4 | E-cdh | 4.00 | 0.0003 |
| 5 | Txnip | 2.54 | 0.0222 |
| 6 | Rab15 | 2.35 | 0.0234 |
| 7 | Dsp | 2.18 | 0.0332 |
| 8 | Grhl2 | 2.17 | 0.0051 |
Additional phenotypes in Grhl21Nisw/1Nisw mutants (but not documented in Rifat et al., 2010 or Werth et al., 2010) include soft tissue syndactyly, lung and heart defects. All five lobes of the lung were present but significantly smaller in Grhl21Nisw versus wildtype, and this difference was clearly evident at E14.5 (Fig. 2S). However, the appearance of the airways at E14.5 and expression of Nkx2.1, a marker of distal lung epithelium, was relatively normal (data not shown). By E18.5 the airways in the mutant lung were significantly smaller than wildtype and they look “collapsed” (Fig. 2T). The Grhl21Nisw heart at E18.5 appears anemic with thinner ventricular walls and larger space within the ventricles (Figs. 2U, V).
Patterning, proliferation and differentiation are not altered in the neural tube of Grhl21Nisw mutants
Drosophila grainyhead acts to repress dorsal, ventral and terminal patterning genes (Biggin and Tjian, 1988; Bray et al., 1989; Huang et al., 1995; Liaw et al., 1995; Soeller et al., 1988), although embryos with a null mutation in grainyhead do not display patterning defects, likely due to maternal provision of grainyhead (Huang et al., 1995; Liaw et al., 1995). To determine whether the neural tube closure phenotype in Grhl21Nisw is due to a patterning defect, molecular markers that reflect patterning along the anterior-posterior and dorsal-ventral axes were examined. Whole embryo or section in situ hybridization on E9.5 wild type and homozygous mutant embryos showed no change in the expression pattern of Shh, Fgf8, Six3, Wnt1, Pax3, Islet1 and Nkx2.2 or in the neural crest marker Sox10 and differentiation at E9.5 was not altered as shown by two early neuronal markers Islet1 and TuJ1 (Supplementary Fig. 2A–S). Proliferation was also not affected as determined by counting the number of phospho-Histone3 positive cells/total cell number at E8.5, E9.5 and E10.5 (Supplementary Fig. 2T; p>0.05).
Expression of genes involved in cell adhesion are affected in Grhl2 mutants
The data above together indicate that Grhl21Nisw/1Nisw mutants cannot be distinguished from wildtype prior to the final step of neural fold fusion and many of the other Grhl21Nisw phenotypes are consistent with defects in tissue closure. This, in combination with the putative role of adhesion molecules in closure events led us to hypothesize that Grhl2 transcription factor controls the expression of one or more adhesion molecules to regulate fusion events in the embryo.
To identify direct and indirect targets of Grhl2, we used microarray analysis to compare gene expression changes between wildtype and mutant embryos. RNA was isolated from the head of E9.5 (23–25 somites) homozygous wildtype and homozygous mutant embryos, at or prior to the time of neural tube closure when the neural folds in wild type and mutant look comparable (formation of hinge points, neural fold elevation, patterning, etc.) and the mutant is determined by genotyping. Affymetrix GeneChip Mouse Exon 1.0 ST microarray experiments and bioinformatic analyses were performed by the University of Colorado Denver Microarray core. A cutoff of 1.25-fold difference was applied resulting in 88 genes downregulated in Grhl2 mutants (p-value <0.04 of which 54 genes had p-values <0.001) and 70 upregulated genes (p-value <0.049 of which 28 genes had p-values <0.01). The full list of genes differentially expressed between mutant and wild type are shown in Supplemental Tables 2 and 3. Quantitative real-time PCR was performed on a subset of these genes and is presented in Table 1.
Of the 40 downregulated genes with the highest fold change in the mutant embryos, 10 encode proteins involved in adhesion (Table 2), with a number of them within the cadherin family of adhesion molecules, including E-cadherin (cadherin-1), cadherin 3, Desmoglein 2 and Desmocolin 2. Desmoplakin was also downregulated in Grhl21Nisw mutants and it is indirectly involved in adhesion by mediating the function of desmosomes. Another group of downregulated genes encode proteins in the Claudin family: Claudin 4, Claudin 6 and Claudin 7. Claudins are components of tight junctions along with occludin and junctional adhesion molecules. Multiple claudins are expressed in tissue-specific manners and these proteins can form homotypic and heterotypic interactions within the tight junction and between adjacent cells to form an adhesive structure (Morin, 2005). Little is known about the expression of claudins during development, thus it is of interest that the microarray data suggests claudins are expressed in the cranial region at E9.5 and their expression is affected by the loss of Grhl2.
Table 2.
Adhesion genes downregulated in the Grhl21Nisw mutant based on microarray analysis
| Gene Symbol | Fold-Change (mutant vs. WT) | p-value (mutant vs. WT) | |
|---|---|---|---|
| 1 | Cldn4 | −2.81 | 3.45E-06 |
| 2 | E-Cdh | −2.76 | 1.75E-05 |
| 3 | Cldn6 | −2.52 | 8.57E-05 |
| 4 | Epcam | −2.51 | 7.64E-06 |
| 5 | Dsp | −2.02 | 1.91E-05 |
| 6 | Cldn7 | −1.68 | 4.73E-06 |
| 7 | Cdh3 | −1.40 | 1.04E-04 |
| 8 | Dsc2 | −1.40 | 2.87E-04 |
| 9 | Dsg2 | −1.33 | 6.41E-04 |
| 10 | Bcam | −1.33 | 1.05E-03 |
The epithelial cell adhesion molecule (Ep-CAM) is a type I transmembrane glycoprotein, with an extracellular domain containing two EGF-like repeats and a short cytoplasmic domain containing two binding sites for α-actinin (Balzar et al., 1999). Ep-CAM can abrogate E-cadherin-mediated cell–cell interactions by disrupting the link between α-catenin and F-actin (Winter et al., 2003). EpCAM can also form complexes with the tight junction protein claudin-7, the variant isoform of cell–matrix adhesion protein CD44v6, and tetraspanin CD9, and this association-facilitated metastasis (Kuhn et al., 2007). BCam, another adhesion molecule (Rahuel et al., 1996), was also downregulated.
Mpzl2/Eva (Epithelial V-like antigen), a new member of the immunoglobulin superfamily (IgSF) (Guttinger et al., 1998), was downregulated. Mpzl2/Eva is expressed in a variety of epithelial tissues and is associated with distinct phases of epithelial differentiation, perhaps through homophilic cell–cell adhesion (DeMonte et al., 2007; Guttinger et al., 1998). Binding interactions between IgSF adhesion molecules are in general weaker compared to the cadherin family, suggesting that IgSF members mediate dynamic intercellular adhesive interactions, rather than strong and static interactions (Guttinger et al., 1998).
The majority of the remaining downregulated genes are expressed, exclusively or largely, in epithelial tissues. For example, Esrp1 is an epithelial cell-type-specific regulator (Warzecha et al., 2009) and keratin8 is found in most extraembryonic and embryonic simple epithelia, including trophectoderm, visceral yolk sac, gastrointestinal tract, lungs, mammary glands, and uterus (Baribault et al., 1993).
Finally, Rax (retina and anterior fold protein) expression was reduced in Grhl2 mutants. Rax is expressed in the anterior neural plate as early as E8.5 and in the optic cup starting at E9.5. Rax null mice have forebrain defects and lack eye structures (Mathers et al., 1997).
The list of upregulated genes was shorter and the fold expression difference less significant. Adamts3 and Adamts1 are members of the Adamts and Adam family of metalloproteases and they can be membrane-anchored (Adam) or secreted (Adamts). Adamts3 can process procollagen II and it is expressed throughout skeletal development and in the hindbrain at E12.5 (Le Goff et al., 2006). Adamts1 is expressed during embryogenesis, predominantly in the epithelium of the lung, pancreas and kidney. Adamts1 null mice are characterized by growth retardation, changes in kidney structure, and impaired female fertility (Shindo et al., 2000). Adamts1 is also upregulated in wounded skin and regulates migration of fibroblasts and endothelial cells (Krampert et al., 2005).
E-cadherin expression is reduced in tissues that express Grhl2
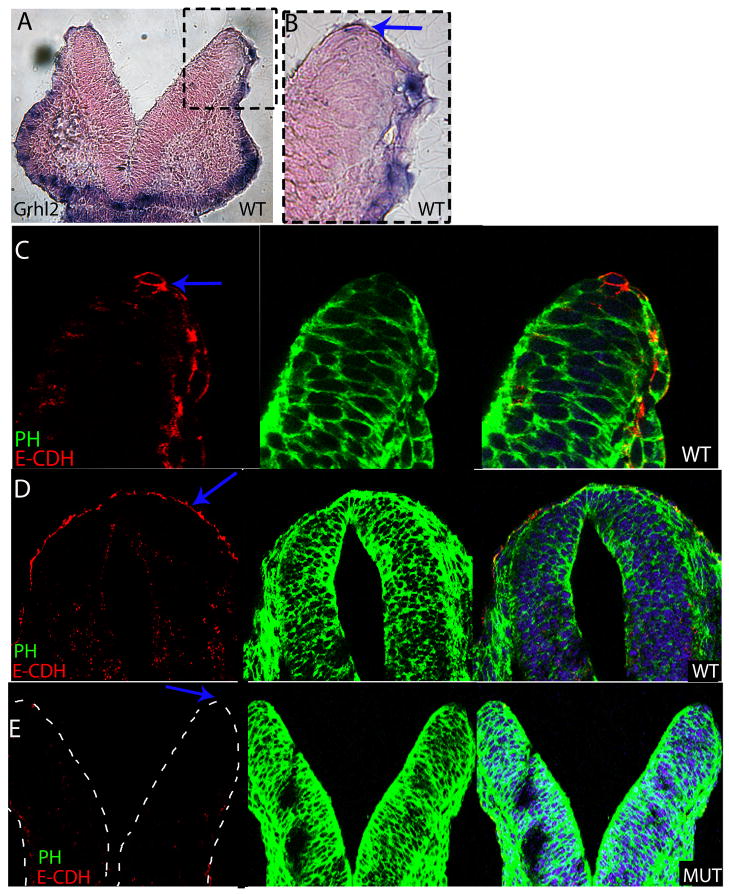
The cadherin family regulates many aspects of development including cell adhesion, delamination and migration. E-cadherin has been hypothesized to be involved in neural tube closure and it was one of the most downregulated genes in our microarray experiment. Therefore, we explored the relationship between Grhl2 and E-cadherin expression in wildtype and mutant embryos. An RNA in situ probe specific for Grhl2 was used to prevent detection of other Grhl family members. At E8.5 Grhl2 is expressed in the ectoderm including the non-neural ectoderm which overlies the neural ectoderm and wraps around the tips of the neural folds as they come into apposition and fuse during neural tube closure (Fig. 3A, B). Grhl3 is also expressed in the non-neural ectoderm during these stages (Auden et al., 2006; Camerer et al., 2010) and Grhl3 function is also required for neural tube closure as Grhl3 null mutants show 100% spina bifida and infrequent exencephaly (Ting et al., 2003a), highlighting the importance of the Grhl genes in neural tube closure in specific rostral-caudal regions (Rifat et al., 2010).
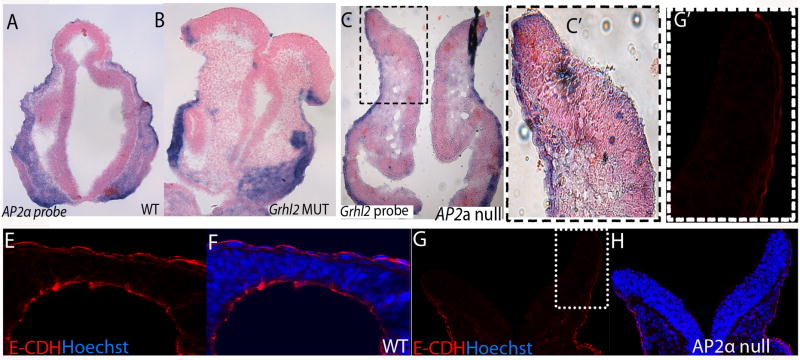
Fig. 3. E-cadherin is downregulated in the neural folds of Grhl21Nisw mutant.
(A) Frontal cryosection of E8.5 wildtype embryo following RNA in situ hybridization with Grhl2 probe. Grhl2 is expressed on the non-neural ectoderm of the neural folds and the craniofacial ectoderm. (B) Detail from panel A showing the non-neural ectoderm of the neural folds.
(C) E-cadherin protein is localized to the non-neural ectoderm of the neural folds at E8.5 in a pattern that corresponds to the expression pattern of Grhl2.
(D, E) E-Cadherin is expressed on the non-neural ectoderm of wildtype embryos (D) at E9.5 whereas there is no detectable E-cadherin expression on the folds of Grhl2 mutants (E). Red is E-cadherin (E-CDH); green phalloidin (PH); blue Hoechst. All frontal sections within the midbrain.
Imaging for A and B: Leica DM 5000B fluorescent microscope and 10X air HC PlanApo objective, NA 0.40. D, E imaged with LSM 510META with 10X air HC PlanApo objective, NA 0.40 and C imaged with 63X oil HCX PL S-Apo objective.
In E8.5 wildtype embryos, E-Cadherin protein is expressed on the non-neural ectoderm and wraps around the neural folds and its localization matches the expression of Grhl2 RNA (Fig. 3C compared to 3B). E-cadherin continues to be expressed in the ectoderm overlying the neural tube at E9.5 and E10.5 (Fig. 3D and not shown). In contrast, in the Grhl2 mutant at E9.5 and E10.5, E-cadherin protein is not detectable in the non-neural ectoderm (Fig. 3E compared to 3D; E-cadherin is also not expressed at E10.5 in the mutant based on microarray and qRT-PCR results). As our recent studies have shown that the non-neural ectoderm initiates fusion of the neural folds (Pyrgaki et al, 2010), our results here combined with the microarray results presented above provide evidence for a role for Grhl2 in the non-neural ectoderm in the regulation of E-cadherin expression, as well as likely other adhesion molecules, in mediating neural fold fusion.
To determine whether Grhl2 is a key regulator of E-cadherin expression in other epithelial tissues, we compared further the expression of Grhl2 and E-cadherin during embryogenesis. In terms of body wall closure and skin development, strong expression of Grhl2 was observed in the developing epidermis at both E14.5 and E18.5 (Supplemental Fig. 3A–D; see also Auden et al. 2006). At E18.5, Grhl2 was specifically expressed on the dermis/epidermis border and the hair follicle (Supplemental Fig. 3C, D). In E18.5 wildtype embryos, E-cadherin was expressed in the epidermis and hair follicles, however, in Grhl2 mutants, E-cadherin expression was not observed in these tissues (Supplemental Fig. 3E, F). There was no obvious effect on follicle shape or structure and β-catenin was normally localized (Supplemental Fig. 3G, H), which was surprising as E-cadherin is thought to be required for β-catenin localization on the plasma membrane.
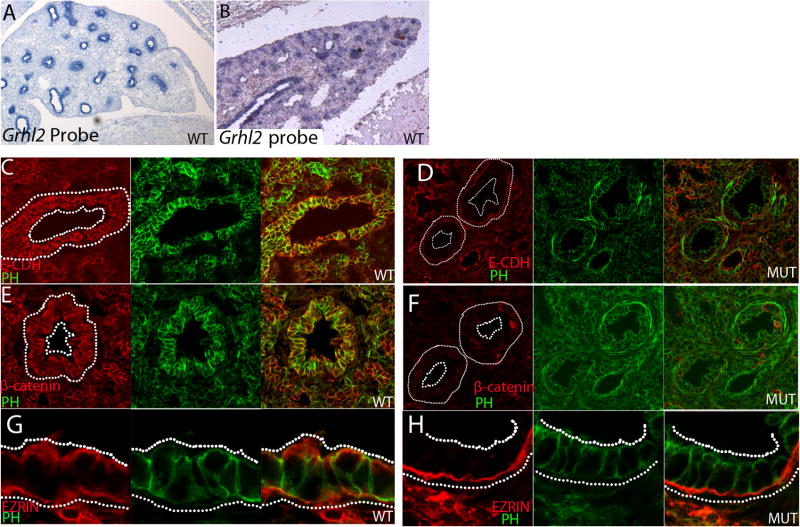
Grhl2 mutants also show a lung defect with smaller airways. Grhl2 RNA was detected in the epithelium of the wildtype lung at E14.5 and E18.5 (Fig. 4A, B respectively) and E-cadherin expression coincided with Grhl2 in the lung epithelium (Fig. 4C). However, in Grhl21Nisw mutants, E-cadherin, as well as β-catenin, expression was severely downregulated compared to wildtype lung epithelium (Fig. 4D, F compared to 4C, E). Ezrin, a known apical marker, was mislocalized and/or downregulated in the epithelium of the mutant lung (Fig. 4G, H). However, the cell shape and overall structure of the epithelial layer was not obviously perturbed in the mutant lung. These data show that Grhl2 in the lung epithelium is required for normal E-cadherin, β-catenin and Ezrin expression and normal lung development.
Fig. 4. Grhl2, E-cadherin, β-catenin and Ezrin expression in the lung epithelium.
(A, B) Grhl2 RNA is detected in the lung epithelium at both E14.5 (A) and E18.5 (B).
(C–H) E18.5 wildtype (C, E, G) and Grhl21Nisw (D, F, H) lungs immunostained with antibodies against E-cadherin (C, D; E-CDH), β–catenin (E, F) or Ezrin (G, H) in red; green is phalloidin (PH). E-cadherin, β–catenin and Ezrin are all downregulated and/or mislocalized in the Grhl2 mutant lung. For A and B: Leica DM 5000B microscope and 10X air HC PlanApo objective, NA 0.40. For C–H Imaging was performed with LSM 510META lens c-Apochromat 40X/1.2 W. C–F 1.3X zoom; G and H 2.2X zoom.
Grhl2 is expressed in many other epithelial tissues in the mouse embryo, as shown here and by Auden et al. (2006). Adhesion of the opposing palatal shelves is critical for palatal fusion and proper face formation and involves several types of cell adhesion receptors. Integrins, cadherin family members, and desmosomal components have been found to be expressed in the prominences during face formation or to cause craniofacial defects when absent (Bader et al., 1998; Ferguson et al., 1992; Frebourg et al., 2006; Mogass et al., 2000; Suzuki et al., 2000). Grhl2 mutants have severe mid-facial clefting and Grhl2 RNA was detected in the facial epithelium at E8.5, E13.5 and E18.5 and on the olfactory epithelium, vomeronasal organ (Jacobson’ organ, E13.5) and serous glands at E13.5 and E18.5 (Fig. 3A, Supplemental Fig. 1H, I). Grhl2 was also expressed strongly in the gut epithelium (Supplemental Fig. 3A).
Embryonic closure is disrupted in AP2α null embryos yet not through a common mechanism of E-cadherin regulation
The AP2α null mouse is one of the few other mouse mutants with severe fusion defects (Zhang et al., 1996), yet the mechanism by which AP2α acts to promote embryonic tissue closure remains unclear. Considering the striking similarity between the closure phenotypes of the Grhl21Nisw and AP2α null mutants (neural tube, body wall, craniofacial), we examined whether there was a relationship between Grhl2, AP2α and E-cadherin. First we investigated whether Grhl2 controls AP2α expression or vice versa. RNA in situ hybridization shows that AP2α is expressed normally in the Grhl21Nisw mutant embryo and Grhl2 is expressed normally in tissues of the AP2α null mouse embryo (Fig. 5A–C and C′). This result correlates with the microarray experiments which showed no change in AP2α expression in Grhl21Nisw mutants. Thus, these molecules do not regulate one another’s expression. We then examined whether AP2α mutants show a change in E-cadherin expression during embryogenesis. By qRT-PCR for E-cadherin RNA in E9.5 wild type and AP2α mutant heads, we found no change in expression (p=0.4) and by immunofluorescence we found E-cadherin protein is expressed and localized in the non-neural ectoderm overlying the neural folds of the AP2α mutant (Fig. 5E–H, G′). This was particularly surprising as previous studies indicated that AP2α binds to the promoter of E-cadherin (Schwartz et al., 2007) and downregulation of E-cadherin expression has been seen upon AP2α disruption in the corneal epithelium and in cancer cell lines (Baldi et al., 2001). Thus, during the critical stage of neural fold fusion, AP2α does not act by regulating E-cadherin protein expression or localization. These data indicate that Grhl2 and AP2α both act to mediate the little understood process of tissue closure, yet they act by different mechanisms. This was further supported by genetic interaction studies in which AP2α+/− heterozygote males were crossed with Grhl21Nisw/+ females and the resulting embryos analyzed. Out of 34 embryos from 4 different litters, 8 embryos were doubly heterozygous, and all of these embryos were indistinguishable from wildtype littermates.
Fig. 5. AP2α null mutation does not result in E-cadherin downregulation.
Frontal sections of E9.5 embryos. (A, B) AP2α RNA is expressed in the neural folds of wildtype (A) and Grhl21Nisw mutant (B) embryos. (C, C′) Grhl2 RNA is expressed in the neural folds of AP2α null mutant embryos.
(E–H) E-cadherin (visualized in red) is expressed in the neural folds of wildtype (E, F) and AP2α null mutant (G, H) E9.5 embryos (blue is Hoechst). G′ detail from G.
For A–C and C′: Imaging with Leica DM 5000B fluorescence microscope, and with 10X air HC PlanApo objective, NA 0.40. For E–H and G′: Imaging was performed with LSM 510META lens c-Apochromat 40X/1.2 W. E–H 1.1X zoom; and G′ 2.2X zoom
Discussion
Neural tube development has been an area of intense research since the beginning of the previous century. A major goal within the field is to relate the knowledge acquired in various animal systems to better understand the causes and methods of prevention of neural tube defects in humans that occur at high frequency and with a significant emotional and medical impact. Despite the substantial increase in knowledge of neural tube development, there are gaps in our understanding of this process. The last step of neural tube formation, joining of the neural folds, is a good example of a poorly understood aspect of neural tube development. Moreover, there are few animal models for the study of closure or fusion of the body wall, optic fissure and face. In this study we explored a novel mutation in mouse that causes a general defect in embryonic tissue closure, affecting closure of the neural tube, face, body wall, and optic fissure.
From a forward genetic screen for neural tube defects, the mutant mouse line, Line2F, was identified and found to have a wide range of phenotypes including a general closure defect as well as abnormalities in lung and heart development. Using a new technology of genomic sequence capture and high-throughput sequencing of the 2.5 Mb region associated with the Line2F phenotype, we identified an ENU-induced point mutation in the Grhl2 gene, a transcription factor that belongs to the Grainyhead-like (Grhl) family of proteins. Mutations in mammalian Grhl2 have been implicated in age-related hearing impairment in humans, although the mechanism is unknown (Peters et al., 2002; Van Laer et al., 2008). In cancer cells, Grhl2 has been implicated in telomerase regulation and cell proliferation (Tanaka et al., 2008; Kang et al., 2009). However, we saw no alteration in proliferation in the embryonic neural tube of Grhl2 mutant mouse embryos. With respect to the function of Grhl2 during embryonic development, our studies and recent results (Rifat et al., 2010; Werth et al. 2010) show that loss of Grhl2 in embryos leads to neural tube defects. In the case of the neural tube, our data indicate that formation of hinge points is not disrupted in Grhl21Nisw mutants and neural fold elevation and apposition occurs normally, and thus we suggest that neural fold fusion is the critical step disrupted in Grhl21Nisw mutants. Our results differ from those of Rifat et al. (2010) in that they report lack of dorsolateral hinge point formation after loss of Grhl2. The discrepancy between their findings and ours may be due to a) the different genetic backgrounds as our results show that Grhl21Nisw phenotypes are strain dependent and b) the use of different mutant alleles. Our work here also extends the role of Grhl2 to show that it is required more generally in the closure of embryonic tissues and development of various organs. Moreover, we show that Grhl2 expression correlates well with the embryonic tissues that show a phenotype in Grhl21Nisw mutant embryos. The discovery of the importance of Grhl2 transcription factor in tissue morphogenesis opens new avenues to explore the cellular and molecular basis of these critical embryonic processes.
In the field of neural tube development, the idea that adhesion molecules are crucial for the process of neural fold fusion has been widely accepted, but very little evidence provides solid support for this idea. For example, Eph5A has been long hypothesized to regulate the last step of neural tube closure, based on its adhesive properties and the fact that Eph5A null mice develop cranial neural tube defects, but definite evidence for such involvement is lacking (Holmberg et al., 2000). The fusion defects in the Grhl21Nisw mutant led us to hypothesize that Grhl2 transcription factor affects the expression of adhesion molecules during embryonic development. To explore this hypothesis we used microarray analysis to identify direct and indirect targets of Grhl2. Our microarray results supports the hypothesis that Grhl2 is involved in regulation of adhesion, as many of the genes that are misexpressed in Grhl21Nisw mutants encode proteins involved in adhesion/epithelial function. We then specifically explored the relationship between Grhl2 and E-cadherin and showed that E-cadherin expression coincides with that of Grhl2 and E-cadherin protein is downregulated in the Grhl21Nisw mutant in the tissues that display a phenotype. The relationship between Grhl2 and E-cadherin was also observed by Werth et al. (2010) and they showed that Grhl2 directly binds to the promoters of E-cadherin and Claudin4 (another target identified in our microarray) to directly regulate their transcription. Our microarray results also reveal a number of other genes affected by the loss of Grhl2. One intriguing possibility is that the Grhl2 transcription factor may bind to the promoter elements of a battery of genes to coordinately regulate the expression of proteins needed for epithelial-mediated morphogenesis. Following this reasoning, the spatial and temporal regulation of Grhl2 expression could induce the expression of a cohort of targets and serve to rapidly alter adhesion and epithelial function in a highly synchronized manner.
One of the top targets of Grhl2 revealed by the microarray experiment was E-cadherin (cadherin1). Homozygous null mutation of E-cadherin is not compatible with life as these embryos show severe abnormalities before implantation (Riethmacher et al., 1995). In the Grhl2 mutant, E-cadherin expression is altered in a specific spatial and temporal pattern. There is a striking correlation between Grhl2 expression and specific downregulation of E-cadherin expression in those tissues that are affected in the Grhl21Nisw mutants (neural tube, face, body wall, skin, lung). In the closing neural folds, Grhl2 and E-cadherin are expressed on the non-neural ectoderm, which is the single cell layer that overlies and wraps around the tip of the neural fold. E-cadherin expression in the non-neural ectoderm is abolished in Grhl21Nisw mutants. Our recent in vivo imaging studies have demonstrated that the non-neural ectoderm initiates the process of neural fold fusion (Pyrgaki et al., 2010). Chen and Hale (1995) cultured rat embryos with an antisense oligonucleotide against E-cadherin and showed that this led to cranial neural tube malformations (Chen and Hales, 1995). Taken together with our results suggesting that fusion is the defective step of neural tube development in the Grhl21Nisw mutants, we propose that disruption of Grhl2 function affects expression of E-cadherin, as well as likely other adhesion molecules revealed by the microarray, causing the non-neural ectoderm to lose its adhesive capacity. This we propose results in failure of the opposing neural folds to adhere, leading to NTD. Although it has been long hypothesized that E-cadherin plays a crucial role in neural tube development, this is the first time that loss of E-cadherin from the non-neural ectoderm of the neural folds is correlated with failure of the neural tube to close. Further studies are necessary to determine whether loss of E-cadherin alone is responsible for NTD. Indeed, our results indicate that other adhesion and apical junctional complex molecules are also downregulated in Grhl2 mutants and therefore, it may be the combined loss of these molecules that disrupts neural tube closure.
E-cadherin also plays an essential role in generation and maintenance of epithelial cell polarity (Behrens et al., 1993; McNeill et al., 1990). An alternate mechanism that could lead to the same NTD outcome is that loss of Grhl2 function, with its consequent effects on E-cadherin and other genes needed for epithelial cell function, may disrupt the structure of the non-neural ectodermal layer so that the cells are misoriented and/or have aberrant polarity and hence cannot associate properly. The epithelium of the neural folds, lung and skin is surprising normal in terms of cell shape and organization. In the mutant lung, there is a loss of E-cadherin, β-catenin, and Ezrin expression, suggesting a defect in cell polarity and/or maintenance of the epithelial cell layer. Indeed, the smaller airways may indicate a structural defect in the lung epithelium. However, this is not a consistent finding as in the skin and hair follicle, despite the loss of E-cadherin, β-catenin is localized normally on the plasma membrane of the cells and appendage structure appears normal.
Our results with the AP2α mutant highlight the molecular complexity of neural fold fusion. Although the closure defects in AP2a null and Grhl21Nisw mutant embryos are strikingly similar, we found that AP2α and Grhl2 do not regulate one another’s transcription and, moreover, AP2α mutants show normal expression of E-cadherin in the neural folds whereas Grhl21Nisw mutants show loss of E-cadherin. Moreover, trans-heterozygotes do not show evidence for genetic interaction. The work described here demonstrates that Grhl2 directly or indirectly regulates an extensive set of genes involved in adhesion and epithelial function, many of which may contribute to the process of neural fold fusion. In the future, it will be of interest to determine whether AP2α regulates a subset of these genes or whether it acts through an independent mechanism. In this respect, recent evidence demonstrates a critical function for protease signaling and Rac1 function from the non-neural ectoderm during neural tube closure (Camerer et al., 2010). Our microarray data on the Grhl2 mutant does not suggest an intersection between Grhl2 and this protease network, at least at a transcriptional level. The present study opens new avenues to explore the commonalities and differences between the molecular pathways regulated by protease signaling, AP2α, and Grhl family members in the closure of embryonic tissues.
Supplementary Material
(A, B) Front view of wild type (WT) and Grhl21Nisw1Nisw mutant (MUT) embryos at E11.5. The mandibular process (m) has closed in both the mutant and wild type embryos. However, the maxillary processes (mx) are widely separated in the mutant, while it has closed in its age-matched littermate. The position of the heart is indicated by H.
(C) Approximately 5% of the Grhl21Nisw1Nisw embryos show a severe phenotype (right) compared to its normal E10.5 littermate (left). The mutant embryos are developmentally delayed, they have not turned and they display an open NT. The position of the heart is indicated by H.
(D, E) Wildtype and Grhl21Nisw mutant eye at E14.5. The optic fissure in the mutant embryo is not closed resulting in a gap in the iris (red arrow).
(F, G) Posterior neuropore (PNP) in E9.5 wildtype (F) and Grhl21Nisw mutant (G). Although there may be a slight developmental delay, the PNP forms normally and we never observe a failure of caudal neural tube closure.
(H, I) Expression of Grhl2 in epithelial tissues at E13.5 (H) and E18.5 (I). Sections through the craniofacial region detected for Grhl2 RNA. At both stages, Grhl2 is expressed in the skin and the olfactory epithelium and serous glands (red arrows).
(A–K) Whole embryo RNA in situ hybridization of E9.5 wildtype and Grhl21Nisw1Nisw mutant embryos shows no change in expression pattern of Shh, Fgf8, Six3, Wnt1 and Sox10. (C–E) Red asterisk: forelimb; green asterisk: midbrain-hindbrain border; purple asterisk: forebrain. (J, K) Red asterisks: Sox10 positive neural crest migrating from the neural tissue towards the branchial arches. (A, B, C, D, J, K) side view; (E, H, I) dorsal view; (F, G) frontal view.
L–O Transverse sections of the rostral spinal cord of E9.5 wildtype (L, N) and mutant (M, O) embryos shows comparable expression of Islet1 and TuJ1 early neuronal markers.
(P–S) In situ hybridization on cryosections of E9.5 wildtype (P, R) and mutant (Q, S) embryos for Pax3 and Nkx2.2, markers of dorsoventral patterning.
(T) Quantification of proliferation as assessed by phospho-histone3 staining in wildtype and mutant embryos at E8.5, E9.5 and E10.5. Each bar indicates the average of the percentage of proliferating cells for all the sections from the embryos of a each embryonic stage (5 mutant and 5 wild type embryos and 6 sections each counted for each age).
(A) Expression of Grhl2 RNA in the developing epidermis including the body wall and gastrointestinal epithelium (GI Ep) at E13.5. The body wall is not fully formed and a portion of the gut still remains outside the body wall cavity. The area denoted by the dotted box is shown in higher magnification in (B) and highlights the epidermis.
(C) Expression of Grhl2 RNA in the skin of E18.5 embryo is limited to the border between dermis and epidermis, and the hair follicle. The box highlights the hair follicle shown in more detail in (D).
(E–H) Cryosections of E18.5 wildtype (E, G) and Grhl21Nisw/1Nisw (F, H) skin visualized with antibody against E-cadherin in (E, F; E-CDH) or β–catenin (G, H) in red; green is phalloidin. (E, F) E-cadherin is observed in the epidermis (EP) and on the outer sheath root of the hair folicle (H) in wildtype (E) but not in Grhl2 mutant (F). (G, H) In both wildltype (G) and mutant (H), β-catenin is present on the membrane of the cells in the epidermis (EP) and in the hair follicle (H). The structure of the epithelial cells and hair follicle is not affected in the Grhl2 mutant, despite the loss of E-cadherin.
For A–D: Leica DM 5000B fluorescent microscope. with 10X air HC PlanApo objective, NA 0.40. (E–H) Imaging was performed with LSM 510META lens c-Apochromat 40X/1.2 W. E–H 2.2X zoom
Acknowledgments
We are grateful to Kathryn Anderson and her lab for help in identifying Line2F, to Ferogh Ahmadi for his efforts in characterizing Line2F phenotypes, to David Pollock, Todd Castoe, and Vijetha Vemulapali for sequencing and data analysis, to Ted Shade and Katerina Kechris for microarray data analysis, to Trevor Williams and Irene Choi for the AP2α mutants, and to the Niswander lab for their insight into this project. LN is an investigator of the Howard Hughes Medical Institute and the work was also supported by NIH R01NS058979 award.
Footnotes
The authors state there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An JY, Kim EA, Jiang Y, Zakrzewska A, Kim DE, Lee MJ, Mook-Jung I, Zhang Y, Kwon YT. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci U S A. 2010;107:1912–1917. doi: 10.1073/pnas.0910267107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auden A, Caddy J, Wilanowski T, Ting SB, Cunningham JM, Jane SM. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns. 2006;6:964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Bader BL, Rayburn H, Crowley D, Hynes RO. Extensive vasculogenesis, angiogenesis, and organogenesis precede lethality in mice lacking all alpha v integrins. Cell. 1998;95:507–519. doi: 10.1016/s0092-8674(00)81618-9. [DOI] [PubMed] [Google Scholar]
- Baldi A, Santini D, Battista T, Dragonetti E, Ferranti G, Petitti T, Groeger AM, Angelini A, Rossiello R, Baldi F, et al. Expression of AP-2 transcription factor and of its downstream target genes c-kit, E-cadherin and p21 in human cutaneous melanoma. J Cell Biochem. 2001;83:364–372. doi: 10.1002/jcb.1235. [DOI] [PubMed] [Google Scholar]
- Balzar M, Winter MJ, de Boer CJ, Litvinov SV. The biology of the 17-1A antigen (Ep-CAM) J Mol Med. 1999;77:699–712. doi: 10.1007/s001099900038. [DOI] [PubMed] [Google Scholar]
- Baribault H, Price J, Miyai K, Oshima RG. Mid-gestational lethality in mice lacking keratin 8. Genes Dev. 1993;7:1191–1202. doi: 10.1101/gad.7.7a.1191. [DOI] [PubMed] [Google Scholar]
- Behrens J, Vakaet L, Friis R, Winterhager E, Van Roy F, Mareel MM, Birchmeier W. Loss of epithelial differentiation and gain of invasiveness correlates with tyrosine phosphorylation of the E-cadherin/beta-catenin complex in cells transformed with a temperature-sensitive v-SRC gene. J Cell Biol. 1993;120:757–766. doi: 10.1083/jcb.120.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Bray SJ, Burke B, Brown NH, Hirsh J. Embryonic expression pattern of a family of Drosophila proteins that interact with a central nervous system regulatory element. Genes Dev. 1989;3:1130–1145. doi: 10.1101/gad.3.8.1130. [DOI] [PubMed] [Google Scholar]
- Brewer S, Williams T. Loss of AP-2alpha impacts multiple aspects of ventral body wall development and closure. Dev Biol. 2004;267:399–417. doi: 10.1016/j.ydbio.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Camerer E, Barker A, Duong DN, Ganesan R, Kataoka H, Cornelissen I, Darragh MR, Hussain A, Zheng YW, Srinivasan Y, et al. Local protease signaling contributes to neural tube closure in the mouse embryo. Dev Cell. 2010;18:25–38. doi: 10.1016/j.devcel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Hales BF. Antisense oligonucleotide down-regulation of E-cadherin in the yolk sac and cranial neural tube malformations. Biol Reprod. 1995;53:1229–1238. doi: 10.1095/biolreprod53.5.1229. [DOI] [PubMed] [Google Scholar]
- DeMonte L, Porcellini S, Tafi E, Sheridan J, Gordon J, Depreter M, Blair N, Panigada M, Sanvito F, Merati B, et al. EVA regulates thymic stromal organisation and early thymocyte development. Biochem Biophys Res Commun. 2007;356:334–340. doi: 10.1016/j.bbrc.2007.02.131. [DOI] [PubMed] [Google Scholar]
- Ferguson MW, Sharpe PM, Thomas BL, Beck F. Differential expression of insulin-like growth factors I and II (IGF I and II), mRNA, peptide and binding protein 1 during mouse palate development: comparison with TGF beta peptide distribution. J Anat. 1992;181(Pt 2):219–238. [PMC free article] [PubMed] [Google Scholar]
- Frebourg T, Oliveira C, Hochain P, Karam R, Manouvrier S, Graziadio C, Vekemans M, Hartmann A, Baert-Desurmont S, Alexandre C, et al. Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J Med Genet. 2006;43:138–142. doi: 10.1136/jmg.2005.031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger M, Sutti F, Panigada M, Porcellini S, Merati B, Mariani M, Teesalu T, Consalez GG, Grassi F. Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis. J Cell Biol. 1998;141:1061–1071. doi: 10.1083/jcb.141.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res A Clin Mol Teratol. 2010;88:653–669. doi: 10.1002/bdra.20676. [DOI] [PubMed] [Google Scholar]
- Holmberg J, Clarke DL, Frisen J. Regulation of repulsion versus adhesion by different splice forms of an Eph receptor. Nature. 2000;408:203–206. doi: 10.1038/35041577. [DOI] [PubMed] [Google Scholar]
- Holmes G, Niswander L. Expression of slit-2 and slit-3 during chick development. Dev Dyn. 2001;222:301–307. doi: 10.1002/dvdy.1182. [DOI] [PubMed] [Google Scholar]
- Huang JD, Dubnicoff T, Liaw GJ, Bai Y, Valentine SA, Shirokawa JM, Lengyel JA, Courey AJ. Binding sites for transcription factor NTF-1/Elf-1 contribute to the ventral repression of decapentaplegic. Genes Dev. 1995;9:3177–3189. doi: 10.1101/gad.9.24.3177. [DOI] [PubMed] [Google Scholar]
- Kang X, Chen W, Kim RH, Kang MK, Park NH. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 2009;28:565–574. doi: 10.1038/onc.2008.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Goodman J, Anderson KV, Niswander L. Phactr4 regulates neural tube and optic fissure closure by controlling PP1-, Rb-, and E2F1-regulated cell-cycle progression. Dev Cell. 2007;13:87–102. doi: 10.1016/j.devcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Kimura T, Takeda S, Sagiya Y, Gotoh M, Nakamura Y, Arakawa H. Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat Genet. 2003;34:440–445. doi: 10.1038/ng1212. [DOI] [PubMed] [Google Scholar]
- Krampert M, Kuenzle S, Thai SN, Lee N, Iruela-Arispe ML, Werner S. ADAMTS1 proteinase is up-regulated in wounded skin and regulates migration of fibroblasts and endothelial cells. J Biol Chem. 2005;280:23844–23852. doi: 10.1074/jbc.M412212200. [DOI] [PubMed] [Google Scholar]
- Kuhn S, Koch M, Nubel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5:553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- Le Goff C, Somerville RP, Kesteloot F, Powell K, Birk DE, Colige AC, Apte SS. Regulation of procollagen amino-propeptide processing during mouse embryogenesis by specialization of homologous ADAMTS proteases: insights on collagen biosynthesis and dermatosparaxis. Development. 2006;133:1587–1596. doi: 10.1242/dev.02308. [DOI] [PubMed] [Google Scholar]
- Liaw GJ, Rudolph KM, Huang JD, Dubnicoff T, Courey AJ, Lengyel JA. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 1995;9:3163–3176. doi: 10.1101/gad.9.24.3163. [DOI] [PubMed] [Google Scholar]
- Liu A, Joyner AL, Turnbull DH. Alteration of limb and brain patterning in early mouse embryos by ultrasound-guided injection of Shh-expressing cells. Mech Dev. 1998;75:107–115. doi: 10.1016/s0925-4773(98)00090-2. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- McNeill H, Ozawa M, Kemler R, Nelson WJ. Novel function of the cell adhesion molecule uvomorulin as an inducer of cell surface polarity. Cell. 1990;62:309–316. doi: 10.1016/0092-8674(90)90368-o. [DOI] [PubMed] [Google Scholar]
- Mogass M, Bringas P, Jr, Shuler CF. Characterization of desmosomal component expression during palatogenesis. Int J Dev Biol. 2000;44:317–322. [PubMed] [Google Scholar]
- Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- Nottoli T, Hagopian-Donaldson S, Zhang J, Perkins A, Williams T. AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. Proc Natl Acad Sci U S A. 1998;95:13714–13719. doi: 10.1073/pnas.95.23.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LM, Anderson DW, Griffith AJ, Grundfast KM, San Agustin TB, Madeo AC, Friedman TB, Morell RJ. Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. Hum Mol Genet. 2002;11:2877–2885. doi: 10.1093/hmg/11.23.2877. [DOI] [PubMed] [Google Scholar]
- Rahuel C, Le Van Kim C, Mattei MG, Cartron JP, Colin Y. A unique gene encodes spliceoforms of the B-cell adhesion molecule cell surface glycoprotein of epithelial cancer and of the Lutheran blood group glycoprotein. Blood. 1996;88:1865–1872. [PubMed] [Google Scholar]
- Rashid D, Newell K, Shama L, Bradley R. A requirement for NF-protocadherin and TAF1/Set in cell adhesion and neural tube formation. Dev Biol. 2006;291:170–181. doi: 10.1016/j.ydbio.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Riethmacher D, Brinkmann V, Birchmeier C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc Natl Acad Sci U S A. 1995;92:855–859. doi: 10.1073/pnas.92.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat Y, Parekh V, Wilanowski T, Hislop NR, Auden A, Ting SB, Cunningham JM, Jane SM. Regional neural tube closure defined by the Grainy head-like transcription factors. Dev Biol. 2010;345:237–245. doi: 10.1016/j.ydbio.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Schwartz B, Melnikova VO, Tellez C, Mourad-Zeidan A, Blehm K, Zhao YJ, McCarty M, Adam L, Bar-Eli M. Loss of AP-2alpha results in deregulation of E-cadherin and MMP-9 and an increase in tumorigenicity of colon cancer cells in vivo. Oncogene. 2007;26:4049–4058. doi: 10.1038/sj.onc.1210193. [DOI] [PubMed] [Google Scholar]
- Shindo T, Kurihara H, Kuno K, Yokoyama H, Wada T, Kurihara Y, Imai T, Wang Y, Ogata M, Nishimatsu H, et al. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest. 2000;105:1345–1352. doi: 10.1172/JCI8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller WC, Poole SJ, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Hu D, Bustos T, Zlotogora J, Richieri-Costa A, Helms JA, Spritz RA. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat Genet. 2000;25:427–430. doi: 10.1038/78119. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kanai F, Tada M, Tateishi R, Sanada M, Nannya Y, Ohta M, Asaoka Y, Seto M, Shiina S, et al. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J Hepatol. 2008;49:746–757. doi: 10.1016/j.jhep.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Ting SB, Wilanowski T, Auden A, Hall M, Voss AK, Thomas T, Parekh V, Cunningham JM, Jane SM. Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nat Med. 2003a;9:1513–1519. doi: 10.1038/nm961. [DOI] [PubMed] [Google Scholar]
- Ting SB, Wilanowski T, Cerruti L, Zhao LL, Cunningham JM, Jane SM. The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem J. 2003b;370:953–962. doi: 10.1042/BJ20021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckfield A, Clouston DR, Wilanowski TM, Zhao LL, Cunningham JM, Jane SM. Binding of the RING polycomb proteins to specific target genes in complex with the grainyhead-like family of developmental transcription factors. Mol Cell Biol. 2002;22:1936–1946. doi: 10.1128/MCB.22.6.1936-1946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laer L, Van Eyken E, Fransen E, Huyghe JR, Topsakal V, Hendrickx JJ, Hannula S, Maki-Torkko E, Jensen M, Demeester K, et al. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum Mol Genet. 2008;17:159–169. doi: 10.1093/hmg/ddm292. [DOI] [PubMed] [Google Scholar]
- Volker JL, Rameh LE, Zhu Q, DeCaprio J, Hansen U. Mitogenic stimulation of resting T cells causes rapid phosphorylation of the transcription factor LSF and increased DNA-binding activity. Genes Dev. 1997;11:1435–1446. doi: 10.1101/gad.11.11.1435. [DOI] [PubMed] [Google Scholar]
- Wan T, Hu Y, Zhang W, Huang A, Yamamura K, Tang H. Changes in liver gene expression of Azin1 knock-out mice. Z Naturforsch C. 2010;65:519–527. doi: 10.1515/znc-2010-7-816. [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Mays JA, Sivak JM, Papagiotas SS, Kim J, Nottoli T, Williams T, Fini ME. Positive influence of AP-2alpha transcription factor on cadherin gene expression and differentiation of the ocular surface. Differentiation. 2003;71:206–216. doi: 10.1046/j.1432-0436.2003.710302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilanowski T, Tuckfield A, Cerruti L, O’Connell S, Saint R, Parekh V, Tao J, Cunningham JM, Jane SM. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech Dev. 2002;114:37–50. doi: 10.1016/s0925-4773(02)00046-1. [DOI] [PubMed] [Google Scholar]
- Winter MJ, Nagelkerken B, Mertens AE, Rees-Bakker HA, Briaire-de Bruijn IH, Litvinov SV. Expression of Ep-CAM shifts the state of cadherin-mediated adhesions from strong to weak. Exp Cell Res. 2003;285:50–58. doi: 10.1016/s0014-4827(02)00045-9. [DOI] [PubMed] [Google Scholar]
- Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:E88–88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- Zhou M, McPherson L, Feng D, Song A, Dong C, Lyu SC, Zhou L, Shi X, Ahn YT, Wang D, et al. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J Immunol. 2007;178:5496–5504. doi: 10.4049/jimmunol.178.9.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Clouston DR, Wang X, Cerruti L, Cunningham JM, Jane SM. Induction of human fetal globin gene expression by a novel erythroid factor, NF-E4. Mol Cell Biol. 2000;20:7662–7672. doi: 10.1128/mcb.20.20.7662-7672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A, B) Front view of wild type (WT) and Grhl21Nisw1Nisw mutant (MUT) embryos at E11.5. The mandibular process (m) has closed in both the mutant and wild type embryos. However, the maxillary processes (mx) are widely separated in the mutant, while it has closed in its age-matched littermate. The position of the heart is indicated by H.
(C) Approximately 5% of the Grhl21Nisw1Nisw embryos show a severe phenotype (right) compared to its normal E10.5 littermate (left). The mutant embryos are developmentally delayed, they have not turned and they display an open NT. The position of the heart is indicated by H.
(D, E) Wildtype and Grhl21Nisw mutant eye at E14.5. The optic fissure in the mutant embryo is not closed resulting in a gap in the iris (red arrow).
(F, G) Posterior neuropore (PNP) in E9.5 wildtype (F) and Grhl21Nisw mutant (G). Although there may be a slight developmental delay, the PNP forms normally and we never observe a failure of caudal neural tube closure.
(H, I) Expression of Grhl2 in epithelial tissues at E13.5 (H) and E18.5 (I). Sections through the craniofacial region detected for Grhl2 RNA. At both stages, Grhl2 is expressed in the skin and the olfactory epithelium and serous glands (red arrows).
(A–K) Whole embryo RNA in situ hybridization of E9.5 wildtype and Grhl21Nisw1Nisw mutant embryos shows no change in expression pattern of Shh, Fgf8, Six3, Wnt1 and Sox10. (C–E) Red asterisk: forelimb; green asterisk: midbrain-hindbrain border; purple asterisk: forebrain. (J, K) Red asterisks: Sox10 positive neural crest migrating from the neural tissue towards the branchial arches. (A, B, C, D, J, K) side view; (E, H, I) dorsal view; (F, G) frontal view.
L–O Transverse sections of the rostral spinal cord of E9.5 wildtype (L, N) and mutant (M, O) embryos shows comparable expression of Islet1 and TuJ1 early neuronal markers.
(P–S) In situ hybridization on cryosections of E9.5 wildtype (P, R) and mutant (Q, S) embryos for Pax3 and Nkx2.2, markers of dorsoventral patterning.
(T) Quantification of proliferation as assessed by phospho-histone3 staining in wildtype and mutant embryos at E8.5, E9.5 and E10.5. Each bar indicates the average of the percentage of proliferating cells for all the sections from the embryos of a each embryonic stage (5 mutant and 5 wild type embryos and 6 sections each counted for each age).
(A) Expression of Grhl2 RNA in the developing epidermis including the body wall and gastrointestinal epithelium (GI Ep) at E13.5. The body wall is not fully formed and a portion of the gut still remains outside the body wall cavity. The area denoted by the dotted box is shown in higher magnification in (B) and highlights the epidermis.
(C) Expression of Grhl2 RNA in the skin of E18.5 embryo is limited to the border between dermis and epidermis, and the hair follicle. The box highlights the hair follicle shown in more detail in (D).
(E–H) Cryosections of E18.5 wildtype (E, G) and Grhl21Nisw/1Nisw (F, H) skin visualized with antibody against E-cadherin in (E, F; E-CDH) or β–catenin (G, H) in red; green is phalloidin. (E, F) E-cadherin is observed in the epidermis (EP) and on the outer sheath root of the hair folicle (H) in wildtype (E) but not in Grhl2 mutant (F). (G, H) In both wildltype (G) and mutant (H), β-catenin is present on the membrane of the cells in the epidermis (EP) and in the hair follicle (H). The structure of the epithelial cells and hair follicle is not affected in the Grhl2 mutant, despite the loss of E-cadherin.
For A–D: Leica DM 5000B fluorescent microscope. with 10X air HC PlanApo objective, NA 0.40. (E–H) Imaging was performed with LSM 510META lens c-Apochromat 40X/1.2 W. E–H 2.2X zoom