SUMMARY
The DNA damage response involves a complex network of processes that detect and repair DNA damage. Here we show that miRNA biogenesis is globally induced upon DNA damage in an ATM-dependent manner. About one fourth of miRNAs are significantly up-regulated after DNA damage, while loss of ATM abolishes their induction. KSRP (KH-type splicing regulatory protein) is a key player that translates DNA damage signaling to miRNA biogenesis. The ATM kinase directly binds to and phosphorylates KSRP, leading to enhanced interaction between KSRP and pri-miRNAs and increased KSRP activity in miRNA processing. Mutations of the ATM phosphorylation sites of KSRP impaired its activity in regulating miRNAs. These findings reveal a mechanism by which DNA damage signaling is linked to miRNA biogenesis.
INTRODUCTION
The maintenance of genome integrity is essential for the proper function and survival of all organisms. Eukaryotic cells have evolved prompt and efficient DNA damage responses to eliminate the detrimental effects of DNA lesions. Upon sensing DNA damage or replication stalling, cell cycle checkpoints are activated to arrest cell cycle progression and allow time for cell to repair DNA damage. The DNA damage response also leads to transcriptional regulation, activation of DNA repair, and when the level of damage is severe, to initiation of apoptosis(Harper and Elledge, 2007). ATM (ataxia-telangiectasia mutated) kinase is one of the key sensors in the DNA damage response, which in particular responds to double-strand DNA breaks (DSB)(Shiloh, 2003).
The ATM kinase preferentially phosphorylates its target proteins at Ser-Gln (SQ) and Thr-Gln (TQ) motifs. ATM-mediated phosphorylation is often associated with increased protein activity or stability(Shiloh, 2001). For example, the checkpoint kinase Chk2 is activated by ATM phosphorylation of Chk2 Thr68 in response to DSB(Matsuoka et al., 2000). ATM and its related kinases, including ATR (ataxia telangiectasia and Rad3 related) and DNA-PKcs (DNA-dependent protein kinase, catalytic subunit), have global effects on many aspects of cellular and organismal function(Durocher and Jackson, 2001). Recent work from the Elledge laboratory unveiled a vast network of over 700 ATM/ATR targets that function in numerous signaling pathways(Matsuoka et al., 2007).
Emerging evidence has shown that microRNAs (miRNAs) are essential modulators of gene expression. MiRNAs are small (~22 nucleotides) noncoding regulatory RNA molecules that are involved in diverse biological processes. They bind to complementary sequences in the 3′ untranslated regions of target mRNA transcripts(Bartel, 2009). MiRNAs are initially transcribed by RNA Polymerase II as primary miRNAs (pri-miRNAs) that include 5′ caps and 3′ poly(A) tails(Lee et al., 2004; Lee et al., 2003). The dsRNA-specific ribonuclease Drosha digests pri-miRNAs in the nucleus to release precursor miRNAs (pre-miRNAs). Pre-miRNAs are approximately 70 nt RNAs with 3′ overhangs, 25–30 base pair stems, and relatively small loops(Lee et al., 2004). Exportin-5, a RanGTP-binding nuclear transporter is responsible for export of pre-miRNAs from nucleus to cytoplasm where Dicer, an endoribonuclease in the RNase III family, cleaves pre-miRNAs to produce mature miRNAs(Chendrimada et al., 2005; Lund et al., 2004).
Several recent reports suggested that miRNA expression is possibly regulated in the DNA damage response. A report from the Persengiev group showed that UV damage triggered relocalization of Argonaute 2 (a key component of RNA-induced silencing complex) into stress granules and promoted miRNA expression in partially ATM/ATR independent manner(Pothof et al., 2009). A subsequent study demonstrated that the tumor suppressor p53 promotes pri-miRNA processing via an RNA helicase p68(Suzuki et al., 2009). MiRNAs also influence the DNA damage response by regulating the expression levels of their target genes. For example, human miR-421 was shown to directly target and suppresses the expression of the ATM transcripts. As a result, overexpression of miR-421 sensitized cells to ionizing radiation(Hu et al., 2010). The human miR-15 and miR-16 cluster targets Cyclin D1 and BCL2, regulating DNA damage-induced cell cycle checkpoints and apoptosis, respectively(Cimmino et al., 2005; Bonci et al., 2008). Our recent study showed that miR-16 is involved in the homeostatic regulation of the DNA damage response through its suppression of the Wip1 phosphatase, a master inhibitor of the ATM-p53 signaling(Lu et al., 2007; Zhang et al., 2010).
In a recent study, the KH-type splicing regulatory protein (KSRP or KHSRP) was identified as a key component in both Drosha and Dicer miRNA-processing complexes. KSRP was originally thought to primarily control mRNA decay(Gherzi et al., 2006; Ruggiero et al., 2007). However, Trabucchi and colleagues provided compelling evidence showing that KSRP also regulates the maturation of a subset of miRNAs, which is potentially required for cell proliferation, apoptosis and differentiation (Trabucchi et al., 2009). In the present study we show that KSRP-dependent miRNAs are included in a class of miRNAs whose expression is induced in an ATM-dependent manner in the DNA damage response. A direct interaction between KSRP and ATM was identified. The ATM kinase directly phosphorylates KSRP, which facilitates the function of KSRP in miRNA maturation.
RESULTS
ATM-mediated DNA Damage Signaling Regulates the Biogenesis of KSRP-associated MiRNAs
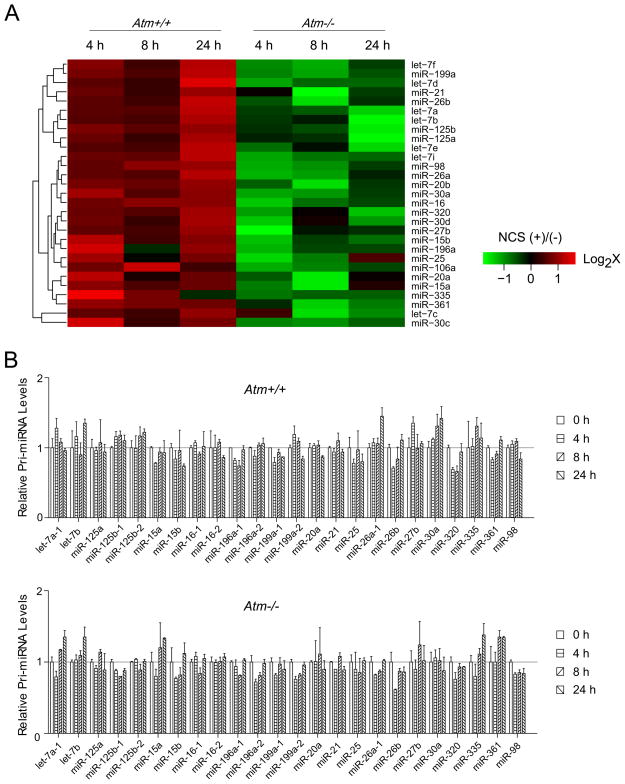
To examine how miRNAs are regulated in the DNA damage response, we assessed the genome-wide mature miRNA expression in Atm+/+ and Atm−/− littermate mouse embryonic fibroblasts (MEFs). MEFs were treated with a radiomimetic drug, neocarzinostatin (NCS) that generates DSBs(Ziv et al., 2006). Cells were harvested at varying time points (0–24 h) and miRNA expression profiling in each sample was determined by mouse miRNA microarray analysis. As many as 71 specific miRNAs, representing about one fourth of total identified mouse miRNAs, were shown to be significantly (≥ 2-fold) induced in the NCS-treated Atm+/+ MEFs, but not in the counterpart Atm−/− MEFs, suggesting that DNA damage stress triggers wide-spectrum alterations on miRNA expression (Figure S1). Comparing these DNA damage-induced miRNAs with the previously reported KSRP-dependent miRNAs(Trabucchi et al., 2009), we found that virtually all of the KSRP-dependent miRNAs (29 miRNAs) were induced in the Atm+/+ MEFs after DNA damage (Figure 1A). More interestingly, the induction of these miRNAs is completely dependent on the functionality of the Atm gene. Loss of ATM in the Atm−/− MEFs completely abolished the DNA damage induction of these miRNAs, suggesting that ATM is probably a pivotal regulator for the activity of KSRP in miRNA biogenesis (Figure 1A). Studies from Trabucchi and colleagues suggested that KSRP is primarily involved in the post-transcriptional processing of miRNAs. To determine whether DNA damage promotes the transcription of these miRNAs, we analyzed the expression levels of primary miRNA transcripts (pri-mRNAs) in both Atm+/+ and Atm−/− MEFs by quantitative reverse-transcriptase PCR (RT-PCR) using primer sets designed specifically for pri-miRNAs. These pri-miRNAs exhibited no significant induction or reduction (< ± 50% change) of their transcription levels during the DNA damage response regardless of the ATM status (Figure 1B). These results suggested that ATM may promote miRNA biogenesis at least in part through KSRP activation.
Figure 1.
DNA Damage Induces the Expression of the KSRP-associated MiRNAs. (A) KSRP-dependent miRNAs are induced after DNA damage in an ATM-dependent manner. Atm+/+ and Atm−/− mouse embryonic fibroblasts were treated with NCS (200 ng/ml), and cells were harvested at indicated time points for microarray analyses. Green or red color on the heat map indicates a decrease or increase of miRNA level and color intensities correspond to relative signal levels on a logarithmic scale. (B) DNA damage has no significant effects on the primary transcript levels of the KSRP-dependent miRNAs. Expression levels of pri-miRNAs in the RNA samples were analyzed by quantitative RT-PCR and error bars represent the mean ± SD.
KSRP-dependent and DNA Damage-induced MiRNA Expression
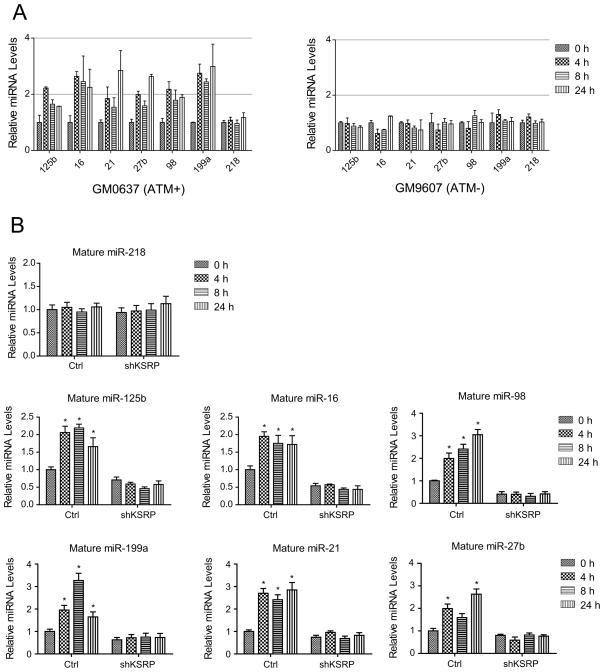
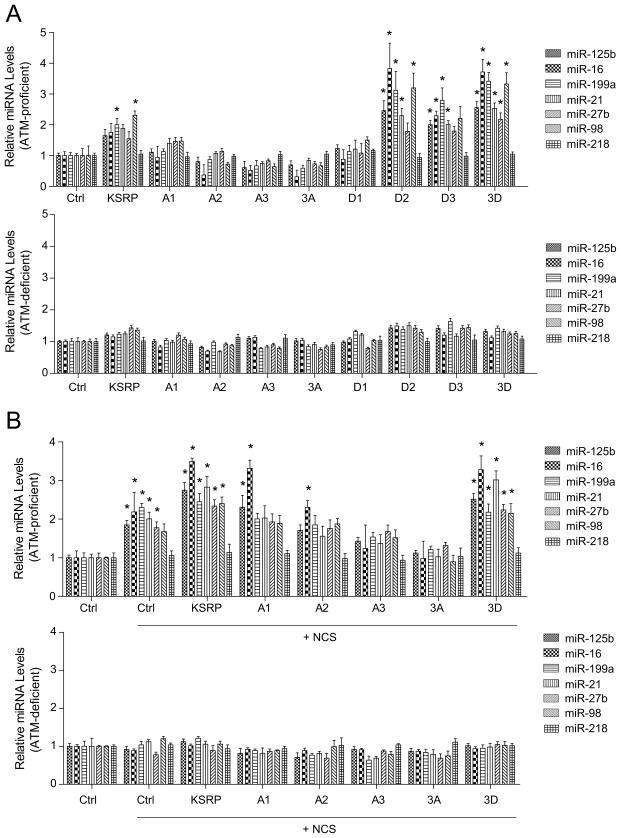
To confirm that the DNA damage-mediated miRNA induction in MEFs was not species-specific, we employed a pair of human fibroblast cell lines with proficient (GM0637) or deficient (GM9607) ATM to assess the levels of six representative mature miRNAs that were randomly selected out of the pool for both ATM- and KSRP- induced miRNAs. The primary transcripts of these miRNAs were not induced after DNA damage in either ATM-proficient or -deficient cells (Figure S2A and S2B. Note that miR-16, miR-125b and miR-199a have two primary transcripts). Consistent with the results from MEFs, the levels of these miRNAs were robustly increased after NCS treatment in ATM-proficient cells, but not in ATM-deficient cells. DNA damage induction was not observed for the control miR-218 that was not regulated by ATM or KSRP (Figure 2A). We further examined whether KSRP is an essential mediator for the ATM-dependent induction of these miRNAs. To this end, U2OS cells stably expressing KSRP shRNA were generated, in which KSRP expression was knocked down by over 80% (Figure S2C and S2D). We assessed the DNA damage induction of the miRNAs in control and KSRP-depleted U2OS cells. Basal levels of mature miRNAs were globally suppressed in the KSRP-depleted cells (Figure 2B). MiRNA induction after DNA damage was dependent on KSRP. Knocking down KSRP clearly abolished the induction of these miRNAs, suggesting a functional correlation between KSRP and ATM in miRNA biogenesis (Figure 2B).
Figure 2.
DNA Damage Induction of miRNAs is Dependent on KSRP. (A) KSRP-dependent miRNAs are induced after DNA damage only in ATM-proficient cells. Human fibroblast GM0637 (ATM-proficient) and GM9607 (ATM-deficient) cells were treated with NCS (500 ng/ml) and mature miRNA levels were determined by quantitative RT-PCR. (B) KSRP-depletion abolishes the DNA damage induction of miRNAs. Control U2OS cells or U2OS cells stably overexpressing KSRP shRNA were treated with NCS and RNA samples were harvested for the expression analyses of mature miRNAs. * P < 0.05, versus the values obtained from the mock-treated controls. Error bars represent the mean ± SD in the figure.
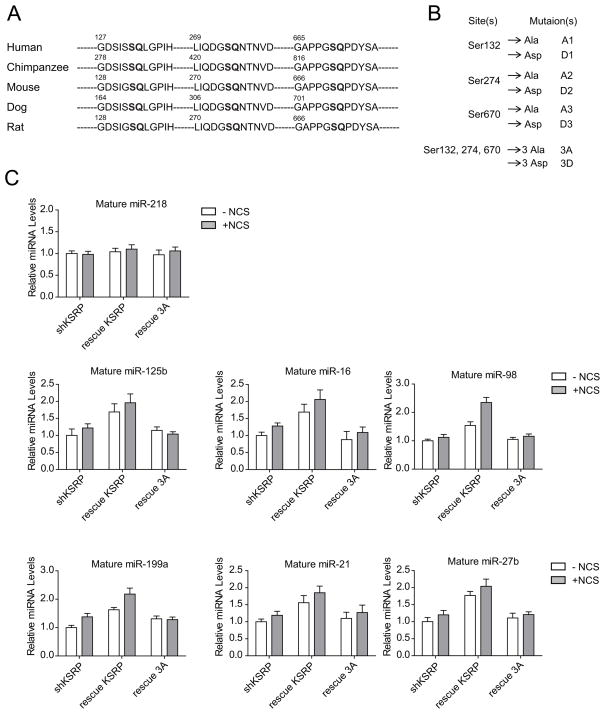
Predicted ATM Phosphorylation Contributes to the Activity of KSRP
The substrates of the ATM kinase have the consensus motif BXBS/TQ, where B residues are often hydrophobic(Kim et al., 1999). Three serine sites (Ser132, Ser274, and Ser670) were identified in a genome-wide search for ATM/ATR targets(Matsuoka et al., 2007). Protein sequence comparisons showed that all of these three sites are conserved in mammals, implicating their functional importance (Figure 3A). To determine if the predicted ATM phosphorylation contributes to the function of KSRP in miRNA biogenesis, we generated a series of mutant forms of KSRP corresponding to each phosphorylation site. Serine to alanine mutation prevents phosphorylation at the mutated site, while serine to aspartic acid mutation mimics constitutive phosphorylation because aspartic acid carries negative charges and structurally resembles phosphorylated serine (Figure 3B). We next tested whether expression of wildtype or mutant KSRP in the KSRP-depleted cells restored the phenotype of DNA damage-induced miRNA biogenesis. Nucleotides targeted by the KSRP shRNA in these KSRP expression constructs were mutated to synonymous ones that encode the same amino acids but are resistant to KSRP shRNA. Overexpression of shRNA-resistant wildtype KSRP greatly increased the levels of the tested 6 mature miRNAs, but not the level of the control miR-218 in untreated cells (Figure 3C, Figure S3A). In contrast, triply 3A mutant of KSRP failed to promote mature miRNA levels. Moreover, inhibiting ATM kinase suppressed miRNA expression even in the absence of DNA damage, indicating basal levels of ATM phosphorylation profoundly contributes to the activity of KSRP in the maintenance of miRNA expression (Figure S3B). Wildtype KSRP-overexpressed cells treated with NCS incurred greater induction of miRNAs than the cells with mock treatment, whereas overexpression of KSRP 3A mutant failed to facilitate the DNA damage induction of miRNAs in the KSRP-depleted cells, highlighting the important roles of ATM phosphorylation in the activity of KSRP (Figure 3C).
Figure 3.
Activity of KSRP in Regulating miRNA Biogenesis is Dependent on the Predicted ATM Phosphorylation. (A) Protein sequence analysis reveals three conserved serines that are potentially phosphorylated by ATM in mammals. (B) A series of point mutations were generated on the predicted phosphorylation sites. (C) Wildtype, but not phosphorylation-deficient mutant KSRP restores DNA damage-induced miRNA biogenesis. KSRP-depleted U2OS cells were transfected with control vector or vector expressing shRNA-resistant wildtype KSRP or 3A mutant of KSRP. Cells were treated with NCS (500 ng/ml) 24 h after transfection and expression levels of mature miRNAs were determined by quantitative RT-PCR. Error bars represent the mean ± SD.
ATM Directly Phosphorylates KSRP
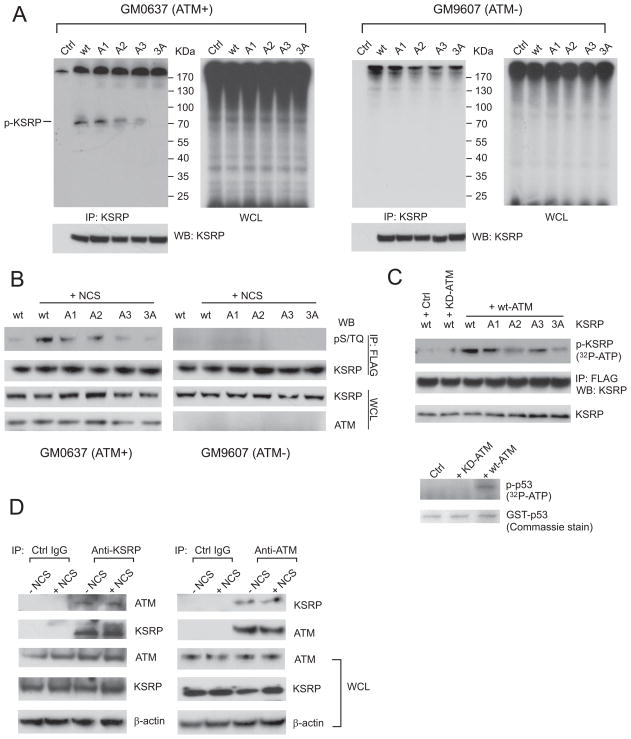
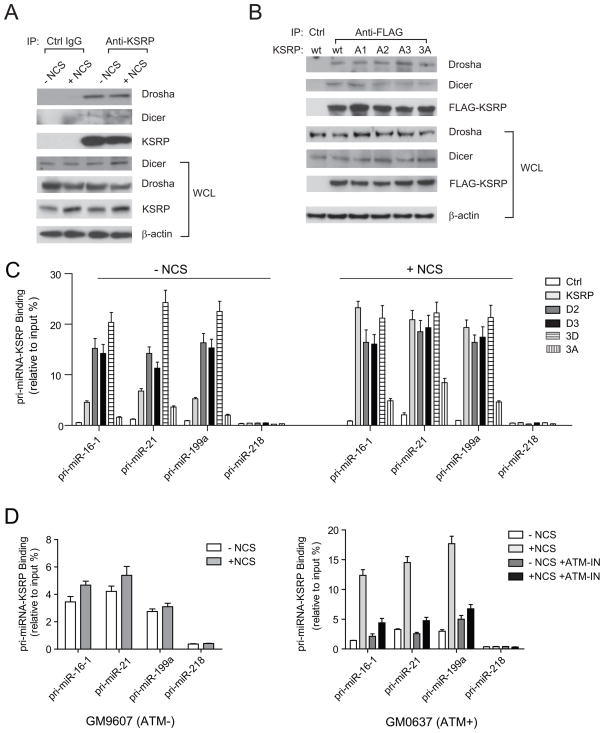
To verify whether ATM directly phosphorylates KSRP, we first performed in vivo phospho-labeling assays by incubating GM0637 cells expressing FLAG-tagged wildtype or mutant KSRP in medium containing 32P-orthophosphate (Figure 4A). Radioactive orthophosphate generates DNA damage in cells without additional DNA damaging agents. In the ATM-proficient GM0637 cells, immunoprecipitated KSRP was shown to be phosphorylated, while mutating each of the three putative phosphorylation sites resulted in diminished phosphorylation levels of KSRP. A2 (S274A) and A3 (S670A) mutations appeared to have greater reduction on the phosphorylation of KSRP compared to A1 (S132A) mutation, indicating that Ser 274 and Ser670 are the major ATM target sites. Due to the ATM deficiency, global protein phosphorylation in whole cell lysates was remarkably impaired in the GM9607 cells and no notable phosphorylated KSRP was detected (Figure 4A). While phospho-specific KSRP antibodies were unavailable, we detected the phosphorylated forms of KSRP in the immunoprecipitated KSRP proteins using a specific antibody that recognizes pS/TQ sites. The results were consistent with the above in vivo phospho-labeling assays, showing the phosphorylation of all three serine sites in ATM-proficient cells, but not in ATM-deficient cells (Figure 4B). The A3 mutation also had the most inhibitory effect on the KSRP phosphorylation (Figure 4B). Because ATM initiates a signaling transduction cascade involving a number of downstream kinases, it is important to know whether the ATM kinase directly phosphorylates KSRP. We incubated purified FLAG-KSRP proteins with immunopurified ATM proteins in the in vitro kinase assays. As a positive control, p53 proteins were phosphorylated by wildtype ATM kinase, but not the kinase-dead mutant (Ser1981 to Ala) ATM (Figure 4C), consistent with the previous studies showing that ATM phosphorylates the Ser15 of p53(Banin et al., 1998; Canman et al., 1998). ATM exhibited robust kinase activity on wildtype KSRP, while phosphorylation levels for mutant forms of KSRP were significantly reduced. A2 and A3 mutations appeared to have greater reduction of ATM phosphorylation, further confirming the above results that these two serines are primary ATM phosphorylation sites (Figure 4C). The triple phosphorylation mutant of KSRP (3A mutant) only showed minimal phosphorylation, meaning that these three predicted sites possibly represent all major ATM target sites (Figure 4C).
Figure 4.
ATM Interacts with and Phosphorylates KSRP in vivo and in vitro. (A) KSRP is phosphorylated in vivo in an ATM-dependent manner. GM0637 and GM9607 cells were transfected with control vector or vector expressing FLAG-tagged wildtype or phosphorylation mutant forms of KSRP. Cells were labeled with 32P-orthophate 24 h after transfection and KSRP proteins were immunoprecipitated by anti-FLAG antibodies. Total phosphorylated proteins or phosphorylated KSRP were analyzed by SDS-PAGE and Western blotting. (B) KSRP is phosphorylated on the predicted ATM target sites. Cells were transfected with wildtype or phosphorylation mutant forms of KSRP and treated with or without NCS. Immunoprecipitated KSRP proteins were analyzed using specific anti-pS/TQ antibodies. (C) KSRP is directly phosphorylated by the ATM kinase in vitro. Purified KSRP proteins were incubated with immunopurified wildtype or kinase-dead mutant ATM in a kinase reaction buffer containing 32P-ATP. Purified GST-p53 is used as a positive control for the kinase activity of ATM. (D) Endogenous KSRP binds to endogenous ATM independently of DNA damage. U2OS cells were treated with or without NCS for 4 h and then lysed for reciprocal immunoprecipitation and Western blotting analyses.
The effects of ATM on KSRP phosphorylation are likely to be direct. We showed that endogenous KSRP binds to endogenous ATM in U2OS cells as measured by reciprocal immunoprecipitation and Western blot analyses (Figure 4D, Figure S4A). DNA damage treatment appeared to have no significant impact on the ATM-KSRP interaction, suggesting that DNA damage-induced protein modifications on ATM or KSRP are not prerequisites for their interaction (Figure 4D). KSRP proteins primarily localize in cell nucleus as shown by immunofluorescence, where ATM proteins are phosphorylated after DNA damage (Figure S4B). A previous study reported that KSRP is involved in the regulation of Dicer and Drosha complex(Trabucchi et al., 2009). Nuclear localization of KSRP may suggest that its primary regulation of miRNAs is associated with the Drosha complex that also localizes and functions in the nucleus.
ATM Phosphorylation Controls the Activity of KSRP on miRNA Induction
To pinpoint the critical phosphorylation site(s) of KSRP, we examined the activity of wildtype or mutant KSRP on the biogenesis of the six miRNAs tested in Figure 2 with or without DNA damage. In the ATM-proficient GM0637 cells, overexpression of wildtype KSRP resulted in ~ 2-fold induction of all of these KSRP-associated miRNAs tested in the experiments (Figure 5A). However, mutating any of the ATM phosphorylation sites markedly impaired the activity of KSRP. Among the three phosphorylation sites, the A1 mutation exhibited less impact on the KSRP activity, while the A2 or A3 mutation greatly reduced KSRP activity. Triple mutation 3A had largely depleted KSRP activity (Figure 5A, Figure S5A). In contrast, D2, D3 and 3D mutations dramatically increased the activation effects of KSRP in miRNA biogenesis, while the effect of D1 mutation was modest. Phosphorylation mutations showed no notable effects on KSRP activity in the ATM-deficient GM9607 cells (Figure 5A). These results strongly suggested that basal levels of ATM phosphorylation contribute to the activation of KSRP in miRNA biogenesis. NCS treatment led to more robust activity of KSRP in the ATM-proficient GM0637 cells, while the A2 and A3 mutations abolished the activity of exogenous KSRP, and triple mutation (3A) even exhibited dominant negative effects compared to the control cells (Figure 5B). Similar to the results from the untreated cells, phosphorylation mutations exhibited no effects in the ATM-deficient GM9607 cells treated with NCS (Figure 5B, Figure S5B). Interestingly, 3D mutant failed to rescue the ATM deficiency in the NCS-treated GM9607 cells, indicating that ATM phosphorylation of KSRP is necessary but not sufficient for miRNA induction after DNA damage. Other ATM targets may also regulate miRNA expression in the DNA damage response.
Figure 5.
ATM Phosphorylation Controls the Activity of KSRP on MiRNA Induction. (A) KSRP induces miRNA biogenesis in the absence of DNA damage. GM0637 (ATM-proficient) and GM9607 (ATM-deficient) cells were transfected with control vector or vector expressing wildtype or mutant forms of KSRP. Expression levels of mature miRNAs were determined by quantitative RT-PCR. (B) KSRP induces miRNA biogenesis in the presence of DNA damage. Cells were transfected as above described and treated with NCS (500 ng/ml) for 4 h. *P < 0.05, versus the values obtained from the control vector transfected samples. Error bars represent the mean ± SD in the figure.
KSRP-pri-miRNA Interaction is Promoted by ATM phosphorylation
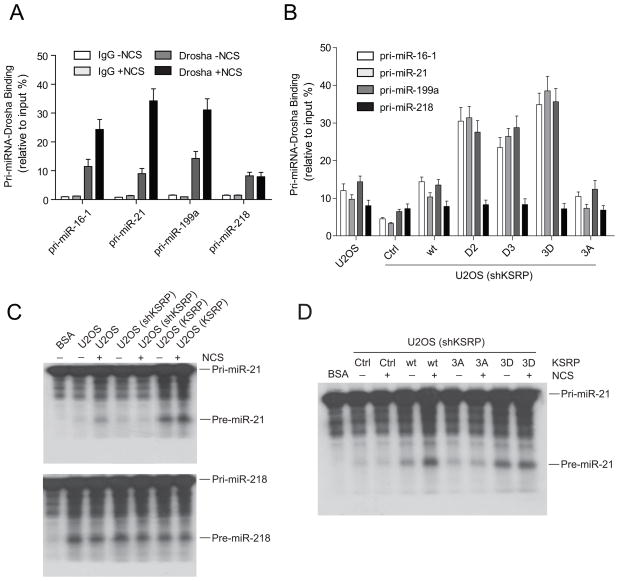
Observations from us and other groups showed that the intracellular localization of KSRP is in the nucleus(Hall et al., 2004; vis-Smyth et al., 1996), suggesting that the primary function of KSRP may involve its regulation of the Drosha complex that processes the primary transcripts of miRNAs in the nucleus. We asked how ATM phosphorylation regulated the function of KSRP in miRNA processing. One possibility is that phosphorylated KSRP binds to the Drosha complex with higher affinity. The interactions between KSRP and Dicer or Drosha were examined by immunoprecipitation and Western blots using NCS-treated or –untreated cells. Both Dicer and Drosha bound to KSRP regardless of DNA damage and DNA damage treatment had no notable effect on the binding capacity of KSRP (Figure 6A). Thus, ATM phosphorylation most probably does not lead to better recruitment of KSRP to the Drosha microprocessors. We also examined whether phosphorylation mutant forms of KSRP retain their binding activity with Dicer and Drosha in the NCS-treated cells. Mutating either one or all of the three ATM phospho-sites had minimal effects on the interaction of KSRP with Drosha and Dicer (Figure 6B). KSRP has been previously shown to bind with a high affinity to the terminal loop of target pri-miRNA and promotes miRNA maturation(Trabucchi et al., 2009). We employed RNA-ChIP (chromatin immunoprecipitation) assays to analyze the interaction of KSRP with pri-miRNAs. We examined the levels of the primary transcripts of three KSRP-induced miRNAs: miR-16-1, miR-21 and miR-199a in the KSRP immunoprecipitates (Figure 6C). In the NCS-untreated cells, about 5.0–7.5% of the three KSRP-associated pri-miRNAs were detected in the KSRP immunoprecipitates. The pri-miRNA binding capacity of KSRP was dramatically increased after DNA damage. Significantly higher percentage (21–25%) of pri-miRNAs was detected in the KSRP immunoprecipitates. As a negative control, pri-miR-218 did not bind to KSRP (Figure 6C). Wildtype KSRP exhibited significantly higher pri-miRNA-binding capacity than phosphorylation-deficient KSRP (3A mutant) in either treated or untreated cells (Figure 6C). D2, D3 and 3D mutations (but not 3A mutation) markedly increased the binding activity of KSRP with pri-miRNAs in the untreated cells, but not in the cells with DNA damage. The results suggested that D mutants functionally mimic the phosphorylated KSRP in the DNA damage response.
Figure 6.
ATM Phosphorylation Enhances the Interaction between KSRP and Pri-miRNAs. (A) KSRP-Dicer and KSRP-Drosha interactions are not enhanced by DNA damage. U2OS cells were treated with or without NCS and cell lysates immunoprecipitated with anti-KSRP antibody. Drosha and Dicer in the KSRP immunoprecipitates were detected by Western blotting. (B) KSRP-Dicer and KSRP-Drosha interactions are not affected by the ATM phosphorylation of KSRP. U2OS cells were transfected with vector DNA expressing FLAG-tagged wildtype or mutant forms of KSRP. Cells were treated with NCS for 4 h. Anti-FLAG antibody was used to pull down FLAG-KSRP proteins. (C) DNA damage promotes the interaction between KSRP and pri-miRNAs. U2OS cells were transfected with control vector or vector expressing FLAG-tagged wildtype or mutant KSRP. Cells were treated with or without NCS. Binding of pri-miRNA to KSRP was determined by RNA-ChIP assays described in the Experimental Procedures. (D) ATM phosphorylation promotes the interaction between KSRP and pri-miRNAs. GM0637 (ATM-proficient) and GM9607 (ATM-deficient) cells were treated with or without NCS (500 ng/ml) in the presence or absence of ATM inhibitor CGK733 (ATM-IN). Binding of pri-miRNA to KSRP was determined by RNA-ChIP assays. Error bars represent the mean ± SD in the figure.
ATM appeared to play a dominating role in the DNA damage-induced KSRP activity, which was examined in two separate contexts: 1) ATM inhibitor (KU55933, 10 μM. Its effects were tested and are shown in Figure S6) dramatically reduced the binding of pri-miRNA to KSRP to a level comparable to that of the untreated samples; 2) NCS treatment failed to promote the pri-miRNA-KSRP interaction in the ATM-deficient cells (Figure 6D).
Phosphorylated KSRP Promotes Pri-miRNA Processing
Pri-miRNAs are processed to pre-miRNAs by Drosha-containing microprocessor. We first explored whether DNA damage induces the interaction of Drosha with pri-miRNAs in RNA-ChIP assays. In the absence of DNA damage, primary transcripts of three ATM-induced miRNAs and control miR-218 bound to Drosha at similar levels (8 to 12% of each total pri-miRNA). While NCS treatment had no detectable effect on the Drosha-binding activity of pri-miR-218, pri-miR-16-1, pri-miR-21 and pri-miR-199a had significantly increased binding with Drosha (2.5–3.2-fold) after DNA damage (Figure 7A). We next examined whether the phosphorylation of KSRP promotes the interaction of pri-miRNAs with Drosha (Figure 7B). Consistent with the previous report(Trabucchi et al., 2009), depletion of KSRP in cells remarkably reduced the binding of the three ATM-induced pri-miRNAs (but not the control pri-miR-218) to Drosha. Reintroducing wildtype KSRP into cells restored the interaction of pri-miRNAs and Drosha. Expression of phosphorylation-mimicking mutants (D2, D3 and 3D) further enhanced the pri-miRNA-Drosha interaction, while phosphorylation-deficient 3A mutant KSRP did not (Figure 7B).
Figure 7.
Phosphorylated KSRP Promotes Pri-miRNA Processing. (A) Pri-miRNA-Drosha interaction is enhanced after DNA damage. Binding of pri-miRNA to Drosha was determined by RNA-ChIP assays using control IgG or Drosha antibody. (B) Exogenous KSRP restores the binding of pri-miRNAs to Drosha in KSRP-depleted cells. KSRP-depleted U2OS cells were transfected with control vector or vector expressing shRNA-resistant wildtype or phosphorylation mutants of KSRP. Binding of pri-miRNA to Drosha was determined by RNA-ChIP assays. (C) KSRP promotes pri-miRNA processing. 32P-labeled pri-miRNAs were incubated with cell extracts from normal and KSRP-depleted or-overexpressed U2OS cells treated with or without NCS (500 ng/ml). (D) Expression of KSRP restores pri-miRNA processing activity in KSRP-depleted cells. KSRP-depleted U2OS cells were transfected with control vector or vector expressing shRNA-resistant wildtype or mutant KSRP. Pri-miRNA processing assays were performed as above described. Error bars represent the mean ± SD in the figure.
We further examined whether enhanced pri-miRNA-Drosha interaction promotes pri-miRNA processing. We performed in vitro pri-miRNA processing assays using 32P-labeled pri-miRNAs and cell extracts from normal and KSRP-depleted or-overexpressed U2OS cells (Figure 7C). NCS treatment significantly increased the pri-miR-21 processing, leaving the control pri-miR-218 processing unaffected. Stable KSRP knockdown reduced the pri-miR-21 processing either in the presence or absence of DNA damage, while overexpression of KSRP in U2OS cells increased both of the basal and DNA damage-induced pri-miR-21 processing activity (upper panel in Figure 7C). The processing activity of the control pri-miR218 was not correlated with KSRP levels and NCS treatment (bottom panel in Figure 7C). We next tested whether addition of wildtype or mutant KSRP restores pri-miRNA processing activity in KSRP-depleted cells (Figure 7D). Expression of shRNA-resistant wildtype KSRP robustly promoted the pri-miR-21 processing and DNA damage further increased the processing activity. While expressing 3A mutant of KSRP exhibited minimal effect, 3D mutant increased the pri-miR-21 processing to a significantly higher level than wildtype KSRP in the cells with DNA damage. The pri-miR-21 processing activity was only slightly induced in the 3D mutant expressing cells after DNA damage, suggesting that 3D mutant is functionally similar to the phosphorylated KSRP after DNA damage.
DISCUSSION
The role of miRNAs in gene regulation is one of the most important and exciting emerging paradigms, linked to multiple facets of cell activities(Bartel, 2009). MiRNA production and maturation are assumed to be regulated by transcription, processing and nucleus-cytoplasm transportation(Davis and Hata, 2009; Siomi and Siomi, 2010). To date, it remains largely unknown whether and how miRNA biogenesis responds to DNA damage, although several recent reports provided evidence indicating that miRNA expression may be modulated in cells treated with DNA damaging agents(Hu et al., 2010; Pothof et al., 2009; Shin et al., 2009; Suzuki et al., 2009; Ishii and Saito, 2006). It is intriguing to hypothesize that primary transcription of miRNAs is probably activated or suppressed upon DNA damage. While genome-wide assessment of the primary transcript levels of miRNAs has yet to be analyzed, we show in the current study that at least for a subset of miRNAs, transcriptional regulation does not appear to account for the DNA damage-induced miRNA expression. Instead, post-transcriptional processing plays a major role in regulating miRNA levels.
As a key kinase to initiate DNA damage signaling cascade, ATM is logically considered to be a potential modulator of miRNA expression in the DNA damage response. In the present paper, our studies started from well-controlled analyses of ATM-dependent alterations of miRNA expression profiling. While a small number of miRNAs had diminished expression in the DNA damage-stressed cells, many more miRNAs were remarkably induced after DNA damage, including miR-16, miR-21 and other miRNAs that are known to be involved in DNA damage checkpoints(Aqeilan et al., 2010; Krichevsky and Gabriely, 2009). We further showed that virtually all of the KSRP-dependent miRNAs were induced after DNA damage. We proposed that ATM-mediated phosphorylation of KSRP may contribute to the induction of these miRNAs. Consistent with this is the fact that ATM directly phosphorylates KSRP, leading to the activation of KSRP and up-regulated miRNA biogenesis. We show that the ATM kinase plays a role in the miRNA processing via substrate phosphorylation. Previous studies have identified two other independent phosphorylation events that involve KSRP-mediated mRNA degradation. Phosphorylation of C-terminal Thr692 by p38 MAPK is essential to regulate the decay of ARE-containing myogenic mRNAs, and Ser193 within the KH1 domain of KSRP is phosphorylated by AKT kinase, which reduces the degradation rate of a subset of mRNAs (az-Moreno et al., 2009; Briata et al., 2005; Garcia-Mayoral et al., 2007). Our results showed that ATM phosphorylation is a major switch for the activity of KSRP in miRNA biogenesis. The activation of KSRP by ATM phosphorylation appears to be site-specific, as phosphorylation on Ser670 and Ser274 imposes more profound effects on the KSRP activity than Ser132 phosphorylation. Protein-binding analyses revealed that the interaction between KSRP and Drosha/Dicer remained intact regardless of DNA damage. However, the binding of KSRP to pri-miRNAs was significantly promoted after DNA damage and phosphorylation-deficient KSRP had severely impaired binding capacity to pri-miRNAs. Ser274 locates in the second KH domain of KSRP and Ser670 is close to the C-terminus(az-Moreno et al., 2009). Although the KH3 domain was considered as a direct binding site for pri-miRNAs, it is reasonable to speculate that phosphorylation events on these two sites probably lead to the conformational changes of KSRP that influence its pre-miRNA binding capacity.
The findings reported here indicate that KSRP plays an integral role in transducing DNA damage signals to the miRNA processing machinery. The effects of ATM on KSRP are multiple and include: (1) facilitation of the KSRP-pri-miRNA interaction, (2) activation of the pri-miRNA processing by Drosha microprocessors, (3) stimulation of miRNA maturation. Through phosphorylating KSRP, ATM integrates a component of miRNA regulatory system into complex DNA damage signaling pathways. In addition to its important functions on mRNA decay, KSRP may act as a molecular gatekeeper that accelerates the production of miRNAs that regulate cell activities in response to DNA damage.
Is phosphorylation of KSRP the primary mechanism by which ATM promotes miRNA expression in the DNA damage response? While we believe this is the case at least for the KSRP-regulated miRNAs, alternative or parallel mechanisms should exist for the rest of DNA damage-induced miRNAs that are not associated with KSRP (Figure S1). Other possibilities include (1) enzymatic activities of Drosha and Dicer may be regulated by post-translational modifications or translocation in the DNA damage response. Multiple phosphorylation sites have been identified or predicted on Drosha and Dicer proteins, some of which may be phosphorylated by ATM or its downstream kinases(Dephoure et al., 2008; Gauci et al., 2009; Brill et al., 2009); (2) transportation of pre-miRNAs from nucleus to cytoplasm is stimulated by DNA damage. Recent proteomic analyses revealed a complex network of Exportin 5-interacting proteins whose levels or binding activity may be altered after DNA damage(Brownawell and Macara, 2002; Chen et al., 2004); (3) transcription of some pri-miRNAs is promoted by activated transcriptional factors. He and colleagues reported that genes encoding miRNAs in the miR-34 family were direct transcriptional targets of p53 and their induction by DNA damage depended on p53(He et al., 2007). A recent study showed that p53 interacts with the Drosha processing complex through the association with DEAD-box RNA helicase p68 and facilitates the processing of several miRNAs (Suzuki et al., 2009).
The ability of KSRP to induce miRNA biogenesis through ATM potentially makes it an important gene in the regulation of tumorigenesis. Homozygous mutations in the ATM gene are responsible for ataxia telangiectasia (AT), a syndrome characterized by acute sensitivity to ionizing radiation and predisposition to cancer. Individuals with ataxia-telangiectasia are estimated to have a 100-fold increased risk of cancer compared with the general population.(Shiloh, 2003; Shiloh, 2001). While only a small population (1%) carries ATM mutations, genes in the ATM signaling pathways are much more frequently mutated in familial and spontaneous human cancers(Shiloh, 2003; Bernstein et al., 2010). Moreover, accumulating evidence from recent studies suggested oncogene-induced DNA damage signaling serve as an anti-cancer barrier in tumorigenesis, underpinning a critical role of ATM in tumor suppression(Bartkova et al., 2006; Bartkova et al., 2005; Halazonetis et al., 2008). Our data clearly showed that loss of ATM abrogated the DNA damage-induced miRNA biogenesis in both human and mouse cells. Some of these miRNAs have been reported to be tumor suppressor candidates, one of which is miR-16. Knockout of miR-16 leads to lymphoid leukemias in mice that are often observed in the A-T patients(Aqeilan et al., 2010). It is predicted that deficient miRNA expression may ultimately contribute to the initiation and progression of tumors. Sood and colleagues recently reported that expression levels of Dicer and Drosha were decreased in 60% and 51% of ovarian-cancer specimens respectively(Merritt et al., 2008). Our preliminary results showed that the expression of KSRP was suppressed in several types of human cancer including cancers in breast, esophagus, kidney, liver and testis (Figure S7), suggesting that KSRP-induced miRNA biogenesis may be inhibited in these cancers.
In summary, we have demonstrated here that ATM plays an important role in the regulation of miRNA biogenesis. ATM directly interacts with and phosphorylates KSRP upon DNA damage, leading to enhanced interaction between KSRP and pri-miRNAs and increased pri-miRNA processing activity by Drosha microprocessors. Our findings on the miRNA regulation in the DNA damage response provides valuable and mechanistic insights into ATM and its associated signaling in suppression of tumorigenesis.
EXPERIMENTAL PROCEDURES
Cell lines and Tissue culture
The U2OS cell line is a human osteosarcoma line that was obtained from the American Type Culture Collection (ATCC). GM0637 (ATM-proficient) and GM9607 (ATM-deficient) are SV40- transformed human fibroblast lines that were obtained from Coriell Cell Repositories. HCT116p53+/+ and HCT116p53−/− cell lines were obtained from the Vogelstein laboratory at Johns Hopkins University. Primary Atm+/+ and Atm−/− mouse embryonic fibroblasts were harvested and cultured as previously described(Lu et al., 2007).
MiRNA Microarray Analysis
Mouse genome-wide miRNA microarray analysis was performed by Phalanx Biotech. The GEO accession number for the microarray data is GSE25916.
Plasmid Constructs and Cell transfection
The FLAG-KSRP expression construct was kindly provided by Dr. Ching-Yi Chen at the University of Alabama. Using this construct, mutagenesis was performed by using QuikChange Lightning Multisite Directed Mutagenesis kit (#210515-5, Stratagene) to obtain constructs expressing mutant forms of KSRP, including A1(S132A), A2(S274A), A3(S670A), 3A(S132A, S274A, S670A), D1(S132D), D2(S274D), D3(S670D), and 3D (S132D, S274D, S670D). ShRNA-resistant KSRP expression constructs were generated from constructs expressing wildtype and mutant forms of KSRP. Anonymous mutations (in parentheses) are shown in the KSRP gene sequence: GAC(T)CAACCGGAGT(A)C(G)CAAGA.
In vivo Phosphorylation Assays
Cells were washed with phosphate-free DMEM (#11971-025, Invitrogen) and subsequently incubated with 0.5 mCi/ml of 32P-orthophosphate in the phosphate-free DMEM containing 10% dialyzed FBS (#26400-036, Invitrogen) for 4 h at 37°C. Cells were harvested and cell lysates were immunoprecipitated by anti-FLAG antibody. KSRP immunoprecipitates were run in SDS-PAGE and then dried for X-ray film exposure(Maya et al., 2001).
RNA-ChIP Analyses
RNA-ChIP was performed as described previously(Trabucchi et al., 2009). Briefly, cells were crosslinked for 20 min with 1% formaldehyde, and cell pellets was resuspended in Buffer B (1% SDS, 10mM EDTA, 50mM Tris-HCl, pH8.1, 1x protease inhibitor, 50U/ml RNase inhibitor). Incubated 10 min in ice, the pellets were disrupted by sonication and the lysates were cleared and subjected to immunoprecipitation with anti-FLAG, anti-KSRP, anti-Drosha, or control antibody, followed by stringent washing, elution and reversal of crosslinking. The RNA was resuspended in 20 μl of TE buffer (10mM Tris-HCl, pH7.5, 1mM EDTA, 50U/ml RNase inhibitor), and incubated with DNase I for 30 min at 37°C to remove any remaining DNA. After extraction with phenol:chloroform:isoamyl alcohol (25:24:1), RNA was precipitated with ethanol and dissolved in 20 μl of DEPC-treated water. 5 μl of RNA was used for the cDNA synthesis reaction. Quantitative PCR reactions were then performed on real-time PCR machine (Realplex2, Eppendorf).
In vitro Pri-miRNA Processing Assays
Pri-miRNA processing assays were performed as described previously(Trabucchi et al., 2009). Briefly, total cell extracts were prepared from U2OS cells and incubated (20 μg per reaction) at 37 °C for 15 min with in vitro synthesized and uniformly labeled pri-miRNAs (5 fmol) in processing buffer containing 100 mM KCH3COOH, 2 mM Mg(CH3COOH)2, 10 mM Tris-Cl, pH 7.6, 2 mM DTT, 10 mM creatine phosphate, 1 μg creatine phosphokinase, 1 mM ATP, 0.4 mM GTP, 0.1 mM spermine, 2 units/μl of RNasin. RNA was purified from the reaction mixture and resolved on 10% polyacrylamide–urea gels that were dried and exposed to X-ray film.
Statistical Analysis
Statistical differences were determined by analysis of variance using one-way ANOVA and Turkey’s Multiple Comparison Test on GraphPad Prism 5 software.
Supplementary Material
Acknowledgments
We thank C. Chen and B. Vogelstein for providing us with KSRP expression vector and HCT116 cell lines. We thank M. Kastan for provision of ATM constructs. We also thank L. A. Donehower for critical discussion and revision on the manuscript. This research was supported by grants to X.L. from the National Institutes of Health (R01CA136549 and R03CA142605) and the American Cancer Society (119135-RSG-10-185-01-TBE), and a grant to F.B. from the National Institutes of Health (P20RR017698).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16–1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- az-Moreno I, Hollingworth D, Frenkiel TA, Kelly G, Martin S, Howell S, Garcia-Mayoral M, Gherzi R, Briata P, Ramos A. Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding. Nat Struct Mol Biol. 2009;16:238–246. doi: 10.1038/nsmb.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bernstein JL, Haile RW, Stovall M, Boice JD, Jr, Shore RE, Langholz B, Thomas DC, Bernstein L, Lynch CF, Olsen JH, Malone KE, Mellemkjaer L, Borresen-Dale AL, Rosenstein BS, Teraoka SN, Diep AT, Smith SA, Capanu M, Reiner AS, Liang X, Gatti RA, Concannon P. Radiation exposure, the ATM Gene, and contralateral breast cancer in the women’s environmental cancer and radiation epidemiology study. J Natl Cancer Inst. 2010;102:475–483. doi: 10.1093/jnci/djq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De MR. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chen T, Brownawell AM, Macara IG. Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol Cell Biol. 2004;24:6608–6619. doi: 10.1128/MCB.24.15.6608-6619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Mayoral MF, Hollingworth D, Masino L, az-Moreno I, Kelly G, Gherzi R, Chou CF, Chen CY, Ramos A. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–498. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81:4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- Gherzi R, Trabucchi M, Ponassi M, Ruggiero T, Corte G, Moroni C, Chen CY, Khabar KS, Andersen JS, Briata P. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 2006;5:e5. doi: 10.1371/journal.pbio.0050005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hall MP, Huang S, Black DL. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol Biol Cell. 2004;15:774–786. doi: 10.1091/mbc.E03-09-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de SE, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci U S A. 2010;107:1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Saito T. Radiation-induced response of micro RNA expression in murine embryonic stem cells. Med Chem. 2006;2:555–563. doi: 10.2174/1573406410602060555. [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, Kastan MB, Katzir E, Oren M. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothof J, Verkaik NS, van IW, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van G, Hoeijmakers JH, Persengiev SP. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero T, Trabucchi M, Ponassi M, Corte G, Chen CY, al-Haj L, Khabar KS, Briata P, Gherzi R. Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling. BMC Mol Biol. 2007;8:28. doi: 10.1186/1471-2199-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, Jin HO, Park IC, Jin YW, An S. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol. 2009;35:81–86. [PubMed] [Google Scholar]
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vis-Smyth T, Duncan RC, Zheng T, Michelotti G, Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem. 1996;271:31679–31687. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70:7176–7186. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.