Abstract
Skin, the largest human organ, is a complex and dynamic ecosystem inhabited by a multitude of microorganisms. Host demographics and genetics, human behavior, local and regional environmental characteristics, and transmission events may all potentially drive human skin microbiota variability, resulting in an alteration of microbial community structure. This alteration may have important consequences regarding health and disease outcomes among individuals. More specifically, certain diversity patterns of human microbiota may be predictive or diagnostic of disease. The purpose of this review is to briefly describe the skin microbiota, outline the potential determining factors driving its variability, posit the likelihood of an association between the resulting microbial community structure on the skin with disease outcomes among individuals, and finally, to present some challenges and implications for studying the skin microbiota.
Keywords: microbial ecology, epidemiology, diversity, transmission, skin
1. Introduction
The skin is the largest human organ. As the skin is in direct contact with the environment, it is inhabited by and constantly exposed to microorganisms in the environment. The resident skin microbiota interacts with other microbes, with human cells, and with the human immune system in multiple ways that mediate risk of disease (Wilson, 205; Wilson, 2008). The purpose of this review is to briefly describe the skin microbiota, outline the potential factors driving its variability, posit the likelihood of an association between the resulting microbial community structure on the skin with disease outcomes among individuals, and finally, to present some challenges and implications for studying the skin microbiota.
For many decades, researchers have been interested in defining the microbial inhabitants of human skin, focusing on descriptive features such as their association with infection (McBride et al, 1977), their stability over time (Evans, 1975), and their interactions with other microbes (Wright and Terry, 1981). Currently, our understanding of the human microbiota is undergoing a dramatic reassessment. The application of high-throughput DNA sequencing to the collection of individual genomes of microorganisms which normally inhabit the human body (the ‘microbiome’) (Peterson et al, 2009) enables characterization of microbial communities in addition to individual microbes. These new studies are using analytic methods from community ecology to describe the structure of the entire microbial community. Community ecology seeks to understand what determines the presence, abundance, and diversity of species in communities, focusing on the role of interactions among multiple species. We are just beginning to use ecological parameters to explore the effects of microbial community structure on disease dynamics within a single host species.
Although there have been some initial reports characterizing the skin microbiota (Dekio et al, 2005; Fierer et al, 2008; Gao et al, 2007; Grice et al, 2008; Grice et al, 2009), most studies to date have focused on the gastrointestinal microbiota. In this system, the role of microbiota diversity in health and disease is unclear. For example, greater fungal richness and diversity were observed in 31 patients with Crohn's disease as well as 26 patients with ulcerative colitis compared to 47 controls (Ott et al, 2009). By contrast, among 3 patients with recurrent antibiotic-associated diarrhea due to Clostridium difficile, bacterial diversity was lower in the fecal microbiome compared to that found among 7 controls (Chang et al, 2008). The sample size of these initial studies are small, and the results do not give a clear picture of whether more or less microbial diversity in the gut is advantageous to the human host. Studies on other microbiota of clinical and general interest, including the oral, urogenital, and skin microbiota (McGuire et al, 2008), also do not show a consistent association between diversity and health and disease. It is still too early to predict whether certain microbial diversity patterns are good or bad, much less whether they cause disease. What is clear is that these patterns are highly complex and dynamic, and require ecological analytic approaches to characterize the microbial communities.
Skin is particularly interesting to study with an ecological approach because of the complexity of its ecosystem. It is composed of an intricate system of cell layers, nerves and glands, protecting the body against extreme environmental conditions, harmful chemicals and pathogens. Keratinocytes, which form the outermost layer of cells on the skin, release antibacterial substances that help prevent infection. Skin also harbors a plethora of different groups of microorganisms that make up the human skin microbiota. Properly characterizing this microbiota has important clinical implications due to its interaction with other microorganisms that may play a role in human disease.
Most studies of skin microbiome have concentrated on characterizing the community structure of microbes inhabiting healthy human hosts or in examining “how particular bacteria become pathogenic” (Chiller et al, 2001; Cogen et al, 2007; Fierer et al, 2008; Gao et al, 2007; Grice et al, 2008; Grice et al, 2009). Though dermatological studies have long since shown associations between a number of skin infections and microbes (Masenga et al, 1990; McBride et al, 1977; Nakabayashi et al, 2000), most have been done using culture-based approaches. Aside from some studies comparing microbial composition between healthy adults and patients with psoriatic lesions (Gao et al, 2008), atopic dermatitis (Dekio et al, 2007), or acne (Bek-Thomsen et al, 2008), there is a surprising lack of literature evaluating potential associations of skin microbiota with health and disease, especially non-dermatological, systemic disease, using molecular approaches. In particular, the role of skin microbiota disturbance on the risk of infectious disease transmission, have not been explored.
Figure 1 describes our conceptual framework for understanding the interactions between skin microbiota, the human host and environment, and the resulting impact on human health outcomes. Significant and potentially harmful alterations of the skin microbial community structure may occur as a result of several factors, including (1) the transmission (dispersal) of non-resident microorganisms into the microbiota, or the removal of dominant microorganisms from the microbiota, both resulting from direct human contact, (2) behavioral characteristics of the individual, such as handwashing practices, (3) local and regional environmental factors, such as the host skin condition and indoor settings, respectively, (4) host genetics, (5) and, host demographic characteristics. Behavioral and environmental characteristics, as well as host genetics and demographics, however, also all have their own direct effects on health outcomes, possibly by affecting host immunity. All of the driving factors included in the conceptual model interact to some degree, as noted by the two-directional arrows (Figure 1). For example, host demographics (e.g. gender) may interact with behavioral characteristics (e.g. cosmetic use) to influence the microbial community structure found on the hands.
Figure 1.
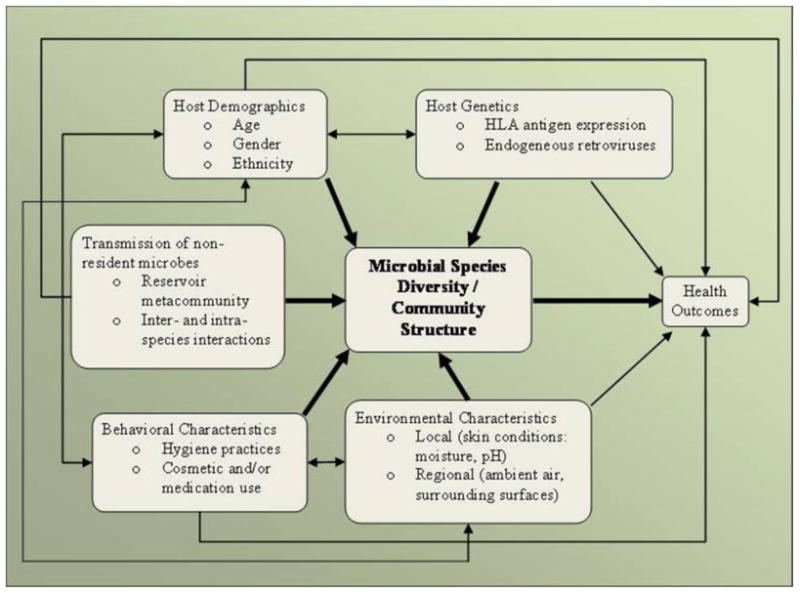
Conceptual framework of the driving forces behind the relationship between microbial species diversity / community structure of the human microbiota and health outcomes. Specific examples are shown as bullet points within each factor.
It is generally accepted that host demographics and genetics, human behavior, certain environmental characteristics, and transmission events can all influence risk of disease. One question that has not been addressed, however, is to what extent these relationships are mediated through the microbial community present on the human body, specifically, the skin. Disturbance of skin microbiota, caused by the various driving factors listed in Figure 1, may influence the course of various disease states.
This review aims to summarize what is currently known about the skin microbiota, the methodological issues regarding how we have come to know it and what needs to be further explored (e.g. temporal dynamics), followed by a summary of each of the determining factors, shown in Figure 1, to be driving human skin microbiota variability.
2. Microbial community structure of human skin
2.1. Skin microbiota
Humans are usually born from an essentially sterile environment (Hrncir et al, 2008; Stecher and Hardt, 2008), but quickly become colonized by microbes. Which microbes become established is primarily driven by the mode of delivery, with vaginally-delivered babies having a microbiota more similar to their mother's vaginal microbiota, and C-section babies having a microbiota more similar to their mother's skin microbiota (Dominguez-Bello et al, 2010). Bacteria and other microorganisms from the environment subsequently interact with the infant's epithelial cells leading to microbial colonization and co-existence. Eventually, an increasingly complex ecosystem forms, comprised of endogenous, or resident, and transient microorganisms (Tlaskalová-Hogenová et al, 2004). These include bacteria, viruses, fungi and protozoa. Humans harbor more microbial cells in their mucosal surfaces and skin than mammalian cells in the entire body (Foxman et al, 2008). While many of them are beneficial, commensal or neutral, some can still become pathogenic (Chiller et al, 2001). It remains to be demonstrated whether the potentially pathogenic members of the microbiota are kept in check by other resident microorganisms. Disruptions by antibiotics, handwashing or lotions may alter the microbial community enabling overgrowth by pathogenic members which then interact with the host causing disease. Additionally, it has been argued that what is considered solely a commensal or a pathogenic organism depends on the profile of the human immune system rather than “the inherent properties of the microbe” itself (Cogen et al, 2007).
The membership of the skin microbiota is quite diverse. A survey of twenty distinct skin sites of ten healthy volunteers using 16S rRNA gene phylotyping, identified 19 phyla and 205 genera (Grice et al, 2009). Using broad-range 16S rRNA genes, PCR-based sequencing of randomly selected clones identified 8 phyla and 91 genera from the superficial volar forearms of six healthy subjects (Gao et al, 2007). In this study, Actinobacteria, Firmicutes, and Proteobacteria accounted for 94.6% of the clones. Using a pyrosequencing-based method, palmar surfaces of the hands of 51 healthy young adult volunteers were surveyed, and shown to harbor more than 25 phyla (Fierer et al, 2008). Of note, the same three phyla accounted for 94% of the sequences in this study. A more comprehensive “whole-body” survey, using a multiplexed barcoded pyrosequencing approach, of 27 body sites (including up to 18 different skin sites) among healthy adults, identified the same three phyla to account for over 82% of the sequences (Costello et al, 2009). According to a recent review of the cutaneous microbiota, “Staphylococcus, Corynebacterium, Propionibacterium, Micrococcus, Streptococcus, Brevibacterium, Acinetobacterium, and Pseudomonas” were named as human skin bacterial residents (Cogen et al, 2007). Many are now emerging as multidrug-resistant pathogens, such as Staphylococcus aureus and Staphylococcus epidermidis. (Marshall et al, 2008; Sommer et al, 2009). Better characterization of the human skin microbiota as “an antibiotic resistance reservoir” has tremendous clinical implications (Sommer et al, 2009).
Most viruses of eukaryotic organisms are not long-term residents on the skin but some “can proliferate within the living epidermis” (Kampf and Kramer, 2004). Only recently has the identification of two commensal viral groups, anelloviruses and GBV-C, alerted attention to the likelihood of a larger human virome (Delwart, 2007). The degree to which other viruses, such as the human papillomavirus and the Merkel cell polyomavirus are endogenous to the skin microbiota, is yet to be fully determined (Singh et al, 2009). Viruses (e.g. hepatitis C virus, rhinovirus, adenovirus, and rotavirus) on the hands have been detected, however, usually as a result of transient hand carriage due to contamination or transmission events (Kampf and Kramer, 2004). Currently, these viral communities are not considered part of the human skin microbiota. The mycological and macroparasitic microbiota of healthy human skin is poorly characterized in comparison to its bacterial and viral counterparts, probably as a result of their rarity and asymptomatic nature (Cogen et al, 2007). Most fungal organisms belong to the genus Malassezia, formerly known as the yeast Pityrosporum (Paulino et al, 2006). Mites, such as Demodex folliculorum, are also “considered part of the normal” microbiota (Fredricks, 2001).
The presence of some microorganisms in the skin microbiota may have an effect on the growth of potential pathogens that may prompt various diseases, indicating the importance of interactions among species (McBride et al, 1977; Selwyn, 1975). For example, sealing certain skin abrasions with band-aids or other hermetic barriers may promote an overgrowth of potentially pathogenic anaerobes, causing a detrimental alteration of the microbiota. S. aureus, once believed to be a “transient colonizer during abnormal conditions”, is now known to be a resident bacterium that somehow turns pathogenic upon disturbance of the individual's skin microbiota (Fredricks, 2001; vanBelkum et al, 2009). Our growing knowledge of skin immunogenetics in the past few years has improved our understanding of the interactions among commensal and potentially pathogenic species (Bowcock and Woodson, 2004; Pivarcsi et al, 2005), however these relationships are not fully understood in terms of microbial communities.
2.2. Methodological Issues in Skin Microbiota Studies
2.2.1. Dependence on sampling methods and laboratory techniques
A representation of the microbiota found on human skin is only as accurate as the sampling methods used to harvest the microorganisms. While swabbing is the most convenient and innocuous of the three, it may not correctly estimate the true microbial diversity across all the skin layers. In comparison, skin scraping picks up more microorganisms per sampled area, but also picks up more skin cells. Punch biopsying, on the other hand, is thought to represent a more comprehensive microbiota since the technique samples across dermal layers (Grice et al, 2008). However, it is the most invasive and covers less surface area in comparison to other methods. Grice and colleagues have compared these sampling methods and concluded that all three yielded the same predominant phylum (i.e. Proteobacteria), and shared over 97% of all bacterial sequences; moreover, all three methods captured very similar bacterial community memberships and structures, as estimated by the high abundance-based Jaccard and Theta (θ) similarity indices, respectively (Grice et al, 2008). In terms of transmission, skin surface sampling of the hands, either by inserting them in a plastic bag filled with buffer solution or by swabbing them, may be the most informative since microbial transmission by humans occurs mostly via direct contact with other individuals and/or environmental surfaces. Estimates of microbial community structure vary by body site sampled. More exposed areas of the body may be composed of “higher proportions of transient microorganisms”, in comparison to lesser exposed areas (Roth and James, 1988). Temperature, moisture level, and amount of sebaceous glands found on the skin vary by body location as well, and may affect where certain microorganisms are found (Grice et al, 2009; Roth and James, 1988).
What is known about human microbiota diversity also depends on the laboratory techniques (i.e. culture-dependent and culture-independent) used to characterize them. The classic approach to identify and quantify microorganisms from the environment has been to culture and differentiate them based on physiological and biochemical tests (Davies et al, 2001; Ogunseitan, 2005). However, culture-dependent methods do not accurately reflect the true bacterial community composition because of the selective properties of the growth media used. Culture-dependent techniques are costly and take time as a result of performing the necessary laboratory tests. After several passages, the microorganisms under study may even behave differently functionally and physiologically. Additionally, some microorganisms will not grow in the absence of others that could be required to provide optimal oxygen, pH, and/or osmotic pressure (Kaeberlein et al, 2002).
Microbial cultivation in the laboratory poorly assess species composition and function in environmental samples. It is generally assumed that “less than 10% of existing microbial diversity in [natural] ecosystems can be accounted for by cultivation” methods (Ogunseitan, 2005). Dekio and colleagues took swab-scrubbed forehead skin samples of five healthy volunteers and analyzed their microbiota using a culture-dependent and a culture-independent method, providing a direct comparison of the two characterization methods (Dekio et al, 2005). Analyses of 16S rRNA gene sequences obtained from the culture-independent method yielded an increased bacterial diversity compared with that derived from the culture methods (Dekio et al, 2005). Culture techniques, therefore, have major limitations in estimating species abundance in natural ecosystems.
Culture-independent microbial DNA-based approaches escape some of these limitations (Ogunseitan, 2005; Theron and Cloete, 2000). Some of these techniques, each having their own relative strengths and limitations, include: 16S rDNA sequencing, PCR and PCR-related techniques, nucleic acid hybridization techniques, polymorphism-based procedures, signature lipid biomarkers, protein profiles, and molecular microarray procedures. All bacteria contain the 16S rRNA gene, which encodes the small subunit of the RNA of the ribosome (i.e. the protein manufacturing machinery of all living cells). It encompasses highly conserved sequence domains interspersed with more variable regions. Identification of bacteria commonly uses the 16S rDNA sequence: conserved regions classify higher taxa, and variable regions differentiate between species. Different variable regions (V1-V9) of the 16S rRNA gene are targeted in different studies of the human skin microbiome, such as the V2 variable region (Costello et al, 2009; Dominguez-Bello et al, 2010; Fierer et al, 2008; Fierer et al, 2010) and the V1-V3 region (Dekio et al, 2005l Dekio et al, 2007). To date, there is no consensus of optimal variable region(s) to target for taxonomic assignment purposes at the genus level or below. Despite the fact that commonly used primers targeting these regions match most of the sequences in most databases, primer biases may still occur where certain phylotypes are missed, thereby generating biased community profiles (Hamady and Knight, 2009).
Though these robust techniques offer a much higher resolution to the characterization of complex microbial communities, they also possess several biases. Nucleic acid extraction and purification, for example, require proper microbial cell lysis and absence of enzymatic inhibitors. PCR techniques can generate artifacts such as chimeras, primer-dimers, and mutations that can lead to a false representation of microbial diversity, and thus require specific and well-designed primers, appropriate nucleic acid starting quantities, and potential PCR elongation time troubleshooting. Another significant limitation to using culture-independent approaches is the inability of the techniques to identify viable microorganisms from samples, leaving unexplained whether the diversity obtained reflects true transients or residents of the skin, or whether they were simply dead contaminants retained on the skin.
The viral portion of the human metagenome, the virome, is more poorly characterized than the bacterial portion. Culture-based approaches suffer from the inability to replicate certain viruses in vitro and the difficulty in establishing viral antigenic/serological cross-reactivity (Delwart, 2007). Culture independent methods, such as shotgun library sequencing and high-throughput pyrosequencing, are also done, though the relatively few numbers of known viral sequences available make it difficult to identify viruses (Delwart, 2007).
2.2.2. Capturing temporal dynamics
Temporal patterns of microbial community structure can be extremely dynamic. Species composition can vary from one time point to another, with irregular cycles. Understanding these patterns is important for investigating associations between the human microbiome variability and health and disease, and ultimately, for determining whether a core human microbiome exists. Identifying a core microbiome for human skin would assist clinical applications, as diagnostic or prognostic factors may depend on recognizing significant deviations from the core. Changes in microbial community structures beyond what is expected over time may indicate an altered physiological state conducive to disease. However, the existence of a core human skin microbiome remains to be determined because at present, the dynamic nature of the skin microbiota has not been adequately characterized.
To date, studies that have characterized the microbial profile of the skin through time, rarely involve more than a handful of intervals (Table 1). Overall, despite the fact that the time variation ranged from a couple hours to ten months in these studies, the skin microbiota was found to be relatively stable. However, these studies are not consistent with each other regarding the health of individuals sampled (e.g. healthy, psoriasis patients, dermatitis patients), the skin site sampled (e.g. facial skin, palmar surfaces), the sampling method (e.g. swab, scrape, scrub-swab), and the method of detection (e.g. pyrosequencing, RFLP analysis, clone libraries) used to characterize their microbiota. Any conclusions about the diversity and/or stability of microbial communities are highly dependent on these sampling issues as well as the taxonomic level analyzed. The inconsistencies between skin microbiota studies make it difficult to generalize results regarding temporal and spatial dynamics of human skin microbial communities.
Table 1.
A summary of selected skin microbiota studies that included temporal dynamics.
| Main Study Aim | Temporal Sampling Done | |
|---|---|---|
| Costello et al, 2009 (Science) | Obtained an integrated view of the spatial and temporal distribution of the human microbiota from up to 27 sites in 7 to 9 healthy adults, using a multiplexed barcoded pyrosequencing approach. | Microbiota samples were donated on 17 and 18 June and 17 and 18 September 2008. |
| Dekio et al, 2007 (J Med Microbiol) | Compared the skin microbiota profiles in 13 patients with atopic dermatitis and 10 healthy controls, using terminal RFLP analysis of bacterial 16S rRNA genes. | Sampled 2 atopic dermatitis patients twice over 7 days. |
| Fierer et al, 2008 (PNAS) | Examined the palmar surfaces of the hands of 51 healthy young adult volunteers to characterize bacterial diversity and to assess its variability within and between individuals, using a novel pyrosequencing-based method. | Swabbed the palms of 4 men and 4 women every 2 h for a 6-h period after hand washing. |
| Gao et al, 2007 (PNAS) | Examined the diversity of the skin biota from the superficial volar forearms of 6 healthy subjects, using 16S rRNA genes PCR-based sequencing of randomly selected clones. | Re-sampled 4 of the 6 subjects 8 or 10 months later. |
| Grice et al, 2009 (Science) | Characterized the topographical and temporal diversity of the human skin microbiome from 20 diverse skin sites of 10 healthy volunteers, using 16S rRNA gene phylotyping. | Collected samples 4 to 6 months after initial visit from 5 of the 10 healthy volunteers. |
| Paulino et al, 2006 (J Clin Microbiol) | Used molecular methods to identify the fungal species present in 25 skin samples from 5 healthy subjects (flexor forearm) and 3 patients with psoriasis. | 2 samples from each forearm of 2 healthy subjects, obtained 10 months apart; 2 samples from same lesion of 1 patient, obtained 6 months apart. |
To obtain a complete understanding of the temporal dynamics of the skin microbiota, it is necessary to capture the community structure at several time points. Figure 2 illustrates the difficulty in determining this dynamic profile. Panels A and B show how sampling (represented by the stars) at two or three different time points may not represent the true variability (represented by the curves) of the microbiota within an individual, and may lead to the conclusion that there is no variability (A) or that some external factor (e.g. treatment) may have contributed to the decrease in microbial diversity in time (B). In reality, the full scope of the variability within an individual can only be determined when sampling is done at sufficient time points (Panel C). Only then, is it possible to speak of an average microbial community structure (represented by the line) per individual (Panel D). Another review recently stated that microbial communities have been thought of as stable because their “temporal variability is lower than inter-individual differences” (Dethlefsen et al, 2006). However, comparisons between the skin microbiota of an individual with another need to be done once the true variability within an individual is known, in order to establish any significant inter-individual differences. Interestingly, by using hierarchical distance-based metrics, Costello and colleagues have shown that human microbial communities cluster first by body site, followed by individuals then time (Costello et al, 2009).
Figure 2.
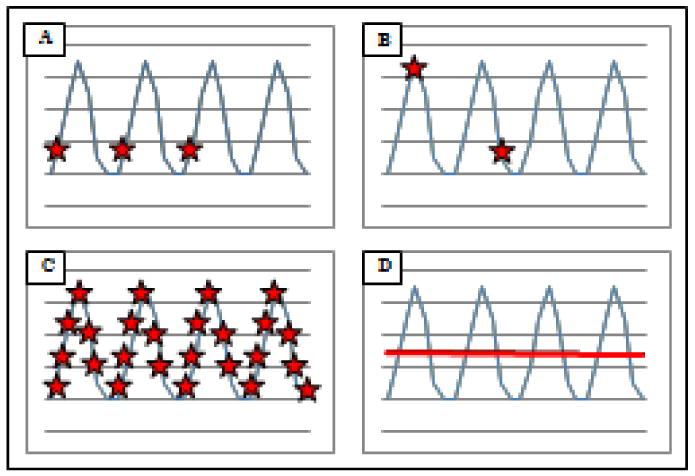
Schematic representations of the potential temporal variability of the human skin microbiome.
To understand the dynamics of the microbial community structure processes, researchers examine the human microbiota over time and space, and the interconnectedness within and between individual hosts, respectively (Foxman et al, 2008). Skin microbial communities have been shown to display specific spatial patterns, with similar communities grouping together at left and right sides of the body, at regions close to the head, and at regions close to the arms (Costello et al, 2009). It has been suggested that, in general, skin sites in closer proximity appear to contain more similar microbial communities than other more distant skin sites (Fierer et al, 2008). Furthermore, certain bacterial phylotypes are shown to predominate moist, sebaceous, and dry skin regions differentially (Grice et al, 2009). Accounting for these spatial differences is important when attempting to capture temporal dynamics among and between individuals. While many studies tend to emphasize spatial and temporal distribution patterns of microorganisms in a specified ecosystem, further explanations of how and why such patterns arise are still largely missing. Human skin microbiota diversity is thought to arise via the many factors depicted in Figure 1 and explored below. It remains to be seen whether any of such drivers of human microbiota diversity act to influence the development of health outcomes.
3. Driving forces of human skin microbiota diversity
3.1. Transmission
Studies that have examined transmission of skin microorganisms often focus specifically on pathogenic microorganisms for the purpose of preventing infectious disease transmission, especially in healthcare settings. Unfortunately, few studies describe transmission of non-pathogenic microorganisms among healthy (and non-healthy) individuals. Additionally, the issues of whether there may be mutualistic relationships between pathogenic and commensal microorganisms that enhance transmission, or antagonistic relationships that minimize acquisition have not been fully assessed. As shown in Figure 1, the role of transmission in influencing the microbial community structure of resident human skin microbiota is very important.
Inter-species interactions can greatly influence the presence of microorganisms within a community. In the skin, for instance, P. acnes and S. aureus have been implicated in working synergistically to increasingly make worse skin lesions caused by one bacterium alone (Lo et al, 2010). Antagonistic interactions also occur, due to competition or predation (Little et al, 2008). For example, S. aureus and S. epidermidis are known to have competitive behaviors on the skin, which could be explained by the serine protease Esp (Iwase et al, 2010) secreted by S. epidermidis, and possibly regulated by its agr pheromones (Otto et al, 2001). As discussed in detail by Chiller and colleagues, “bacteriocin and toxic metabolite production, induction of low reduction-oxidation potential, nutrient depletion, and inhibition of adherence and translocation” are just a few of the mechanisms used by bacteria that allow them to interact in the same community (Chiller et al, 2001). For example, bacteriocins, which are toxins produced by certain bacteria (e.g. lactobacilli, propionibacteria), are able to inhibit the growth of other, potentiallymore pathogenic bacteria (e.g. staphylococci) (Klaenhammer, 1993; Oh et al, 2006). Among those with damaged skin, certain bacteriocin producers proliferate and dominate the bacterial community (Roth and James, 1988). Novel bacteriocins are being identified at a growing pace (Martin-Visscher et al, 2008; Sawa et al, 2009; Tiwari and Srivastava, 2008). The bacteriocin nisin, from Lactococcus lactis, has been shown to reduce the clinical signs of mastitis, which is generally caused by a Staphylococcal infection (Fernandez et al, 2008). Indirect inter-species interactions also occur through the engagement of the host immune system (Chiller et al, 2001). Viral infection, for example, causes alterations on epithelial cell surface receptors (Roth and James, 1988).
Transmission via direct contact with other individuals or indirectly with fomites or water droplets found in the environment introduces transient microorganisms that may have the potential to alter the dynamics between resident microorganisms of the skin microbiota. Moreover, movement patterns of daily living may have an effect on microbiota, in its ability to enhance transmission probabilities. For instance, the number of people living in close contact with an individual and their networking patterns, the individual's commuting practices, occupation and leisure pursuits, and interchanges with schools or child-care facilities all could influence the spread of skin microbes.
3.2. Host demographic characteristics
The microbial communities present on skin are determined by skin conditions, the host's hormonal status, age, gender, and ethnicity (Figure 1) (Fierer et al, 2008; Fredricks, 2001; Grice et al, 2009; Roth and James, 1988). In terms of skin conditions, overall the skin is cooler than core body temperature, and has a pH around 5, although it varies by body site (Chiller et al, 2001). Several molecules synthesized by the skin can contribute to skin surface conditions, which for the most part have the ability to discourage microbial growth. Even host gender shapes skin environment, thereby influencing what is able to colonize men and women. Women have been shown to have significantly greater bacterial diversity on their hands in comparison to men (Fierer et al, 2008). Though there have been reports of differences in carriage rates of microorganisms between races, there is still much to learn regarding the diversity of microbiota across a wide range of cultures and ethnicities (Evans et al, 1984; Mai and Draganov, 2009; Sultana et al, 2003). Explanations for age and gender related differences may include differences in hormones, sweat or sebum production, skin pH differences, and interactions with host behavior. For example, a plausible explanation for women having greater bacterial diversity on the skin of their hands may be that they likely have more contact with children, who commonly experience a high burden of common infectious diseases. Also, women may be more likely to use cosmetics, thereby altering the microbial community structure of their skin. A survey looking at potential associations between demographic information of neonatal intensive care unit nurses and the total microbial composition found on their hands, showed that while age had a minimal effect, race was shown to be “a significant predictor of skin health” (Sultana et al, 2003). The authors note, however, the limited variation in age among the nurses surveyed.
3.3. Host genetics
Apart from skin's structural cell layers and synthesis of molecules that influence microorganism proliferation, its immunological machinery may also play a role in skin microbial community structure. The innate immune system of the skin, now known to be important in regulating the microbiota at multiple epithelial surfaces, contains Langerhans cells, T lymphocytes, mast cells, and keratinocytes, which expresses Toll-like receptors and produces cytokines, chemokines, β-defensins, RNase7, and other antimicrobial peptides (Pivarcsi et al, 2005). The skin-associated lymphoid tissue (SALT) has the ability to produce and secrete immunoglobulins, present antigens and activate T-cells, which can affect the composition of the microbial communities. Conversely, certain skin microbial residents are known to affect the host immune system. For instance, S. epidermidis has recently been shown to upregulate the expression of antimicrobial peptides in keratinocytes (Wanke et al, 2010). Immunogenetic components of the skin, such as the human leukocyte antigen (HLA) gene cluster, are shown to have associations with certain skin diseases, such as psoriasis (Bowcock and Woodson, 2004) and ashy dermatosis (Correa et al, 2007). It remains to be shown whether such associations are mediated by the microbial communities of the skin (Figure 1).
While genome-wide association studies (GWAS) have successfully identified many genetic variants to be associated with a number of human diseases, GWAS investigating whether genetic variations in the human genome can influence microbiota composition are now emerging, owing in part to rapid advancements in sequencing technologies and bioinformatics. Commonly measured genetic variants include single nucleotide polymorphisms (SNP), non-SNP variants, and insertion-deletions (INDEL). Using a large murine intercross population, Benson and colleagues were able to show that host genotype does indeed explain some of the variation in the gut microbiota, controlling for environmental factors (Benson et al, 2010). Another murine model study looked for associations between the matriptase genetic variant, which led to filaggrin deficiency and atopic dermatitis phenotype, and skin microbiota (Scharschmidt et al, 2009). Scharschmidt and colleagues were able to show a significant alteration in the skin microbiota of these transgenic mice, in particular, a higher abundance of Corynebacteria and Streptococci, in comparison to their wild-type littermates (Scharschmidt et al, 2009).
3.4. Human behavioral characteristics
Behavioral factors such as the use of medications (e.g. antibiotics, steroids), hygiene practices (e.g. personal, domestic), and use of cosmetics (e.g. creams, lotions, emollients) have all been reported as having the ability to alter the microbial community structure of the skin (Figure 1) (Fierer et al, 2008; Fredricks, 2001; Grice et al, 2009; Larson, 2001; Larson et al, 2002; Roth and James, 1988). Other behavioral characteristics such as diet and nutrition, sun exposure, and smoking, are all considered contributing factors to skin and systemic health, however, their potential to influence the microbial community structure of skin has yet to be examined.
Hand washing has been long been considered to be the simplest and most effective method for controlling infectious diseases (Borges et al, 2007; Larson, 2001). Individuals' hands can be thought of as either fomites, by transiently carrying microorganisms, and/or as vectors, by harboring established, endogenous microorganisms that have the potential to be transmitted from one person to another. Despite the multitude of studies emphasizing the benefits of personal hygiene on reducing disease transmission by removing transients obtained by contamination (Aiello et al, 2008; Allengranzi and Pittet, 2009; Larson, 2001; Larson et al, 2004; Luby et al, 2005), the effects of hand washing on the microbial community structure of the hands is an area in need of more research. We still do not know the impact of hand washing on the longer term resident biota. However, such impacts have already been metaphorically equated to the disturbance caused by “hurricanes” and “forest wildfires” (Fredricks, 2001; Marris, 2009). Most reports of resulting microbial structure alterations rely on total bacterial colony-forming-units (CFU) in an attempt to explain disease causality. However, although hand washing is meant to remove transient microorganisms to either decrease self-inoculation (why we wash hands before eating) and/or transmission (why we wash hands after sneezing into them), researchers do not necessarily see a reduction in CFU counts after hand washing (Aiello et al, 2003). This may be a consequence of a microbial community structure disturbance whereby shedding of the skin reveals another layer of resident microorganisms. Realistically, diseases occur not just with an increase in bacterial loads, but also with an alteration in the microbiota of the individual and the resulting interaction with host immunity. Aside from reducing the number of transient microorganisms present on the skin, hand washing also has an impact on the skin condition itself, in altering the resistance capacity of the stratum corneum (i.e. the electrical properties of the skin), lipids, transepidermal water loss, and pH, which could consequently affect the microbial community structure (Larson, 2001). Hand washing can be seen as a disturbance to the microbial community structure, possibly perturbing the existing trade-off between its microbial colonizers and competitors.
Individuals differ widely in their behavioral habits, which may have potentially meaningful consequences in altering the skin microbiota. Just in terms of hand hygiene alone, the frequency and duration of washes and type of soap product use (plain soap, antimicrobial soap, and/or alcohol sanitizers), can account for some of the variation in microbial community structure between individuals
3.5. Environmental characteristics
Temperature, moisture, and exposure to ultraviolet radiation, are all known examples of environmental factors that can alter skin conditions and have the potential to influence the microbial community structure of the skin (Figure 1). For colonization to take place, microorganisms must adhere to a host by binding to specific receptors on the host epithelial cell, and have been shown to do so with varying affinities (Romero-Steiner et al, 1990). Though skin dryness may help to prevent the acquisition of certain transient microorganisms, consequent breaks on the skin surface may expose such receptors (Roth and James, 1988). Seasonality has been demonstrated in influencing diseases of the skin, likely as a result of microbiota alterations in response to climate changes (Jha and Gurung, 2006). Moreover, ultraviolet B (UVB) radiation, known to impact skin conditions, was observed to have disparate microbicidal effects on the skin microbiota (Dotterud et al, 2008). In particular, S. aureus appeared to be more sensitive to the radiation treatment than S. epidermidis.
Individuals are constantly being exposed to the microbial fluctuations of the indoor environment (Rintala et al, 2008). In addition, individuals differ widely in their occupational exposures (e.g. nurses, gardeners, teachers), which may also account for the variation between the community structures of their skin microbiota. For example, significant skin microbiota differences were observed between chronically ill outpatients and hospitalized inpatients, controlling for chronic illness as a potential confounder, which may indicate hospitalization as a potential driver of variability (Larson et al, 2000).
Just as the skin microbiota of humans harbor resident microorganisms, the physical environment (e.g. door handles, kitchen surfaces) surrounding the individual may be “reservoirs” for microbial colonization (Kagan et al, 2002). Even house dust has its own characteristic microbial composition (Maier et al, 2010; Rintala et al, 2008). Microbial communities within showerhead biofilms across the United States were found to contain opportunistic human pathogens (Feazal et al, 2009). These potential reservoirs likely increase the risk of microbial transmission, and thus the opportunity for disease. Concerns about the impact of environmental determinants of health are important, but their influence in altering the microbial community structure of skin microbiota, thereby resulting in adverse health outcomes, has not been sufficiently investigated. The fact remains that not much is known about the microbial composition of environmental settings (Feazel et al, 2009), nor how it influences the microbial community structure of skin.
3.6. The impact of microbial community structure on health outcomes
Underlying biological mechanisms explaining why an altered skin microbiota diversity may result in disease, thus explaining the arrow from ‘species diversity / microbial community structure’ to ‘health outcomes’ in Figure 1, include inflammation, absence of necessary members of the microbial community, and a decrease in microbial antagonistic interactions (Stecher and Hardt, 2008). Other possible mechanisms may consist of modifications to normal microbial signal transduction and quorum-sensing, resulting in cascades that may lead to damaging cellular changes in the host. Lateral gene transfer may allow skin microbiota to share functional roles, possibly eliminating redundant species and consequently impacting host health outcomes.
Given the many different ways in which the skin microbial community structure can be modified to potentially play a role in disease, it is clear that it is not the mere acquisition of a pathogen that causes disease, or hand washing that directly prevents disease, or that contaminated surfaces result in disease, or even that antibiotics eliminate disease. These events are all mediated by the resident skin microbial community structure of the individual. It is what happens to that microbial diversity that governs whether or not a host immune response is elicited, thereby establishing disease. In demonstrating that the skin microbiota is responsible for controlling cutaneous inflammatory responses, thereby protecting the host from unintended inflammatory diseases, Lai and colleagues provided evidence of a relationship between the microbiota, host immunity, and disease (Lai et al, 2009).
The microbial diversity present in and on humans is associated with several infectious and non-communicable diseases. Changes in resident microbial communities have been shown to be associated with skin conditions such as acne, atopic dermatitis, and psoriasis (Bek-Thomsen et al, 2008; Dekio et al, 2007; Gao et al, 2008). Even more broadly, The Human Microbiome Project has inspired exciting new studies demonstrating how changes in resident microbial communities play a role in disease, including antibiotic-associated diarrhea, bacterial vaginosis, human immunodeficiency virus, obesity and cardiovascular disease (Oakley et al, 2008; Ordovas and Mooser, 2006; Othman et al, 2008; Price et al, 2010; Young et al, 2008). Insights into the effects of resident microbial diversity of the human microbiota on health outcomes provide encouragement for further characterization of the skin microbiota.
4. Conclusions
4.1. Challenges in assessing multilevel associations
Describing associations between the microbial community structure found on the skin and the health outcomes of individuals requires an integrative approach across several disciplines. Studying the human microbiota involves the fields of microbial ecology, population biology and microbiology. Further linking the skin microbiota to individual and population health outcomes also incorporates medicine, immunology, epidemiology and biostatistics. Thus, a comprehensive understanding of correlations between changes in the human microbiota and disease, with the consequent translation into public health benefits, requires an interdisciplinary endeavor.
Without an understanding of the normal range of microbial diversity within and between individual hosts, it is difficult to relate microbiota composition to disease status (Mai and Draganov, 2009). Further complication arises from recognizing that microbial diversity involves many levels: the microbial level (individual microbes as well as populations and communities of microbes), the individual level (host factors), and the human population level. Despite a number of ecological studies that assess population-level risk factors for disease, most epidemiological studies have traditionally looked at individual-level risk factors (Diez Roux and Aiello, 2005). Recently, however, it has become more apparent that focusing on the individual level does not account for other equally important health determinants such as the influence of social norms (a population level factor), like hygiene practices, on disease risk (Larson et al, 2004).
In any attempt to infer a causal association between human microbiota and disease, it is necessary to determine the risk of developing disease given the present microbial community on a host population. Therefore, another challenge, one effectively explained by Mai and Draganov, is the need for longitudinal studies with enough power to identify microbiota differences between groups despite the large variation that is likely to be observed within groups (Mai and Draganov, 2009).
4.2. Implications for Health
In this review, we have shown that skin, the largest organ of the human body, is normally colonized by a diverse community of microorganisms, some of which are potentially pathogenic under certain conditions. It is the continuing inter- and intra-species interactions of the microbial community, along with host immunity, that regulate these conditions to avoid disease. An implication of this regulation is that when microbial community interactions are altered, certain microorganisms may become more easily dispersed and thus be more readily transmitted to another person or even oneself (i.e. autoinfection). Additionally, keeping the skin microbiota in check may allow the host immunity to be continually primed, so that in the event of disease onset, it is better equipped at controlling its progression.
We have also shown that the microbial community structure of the human skin is continuously influenced by microorganism dispersal, host behavioral characteristics, and the environment. These driving factors may lead to significant and potentially harmful alterations of the skin microbiota. The implication is that by manipulating the human skin microbiota community structure via its modifiable transmission-related, behavioral, and environmental pathways (Figure 1), disease could potentially be prevented or treated, especially given the recent advances in molecular technology. The most obvious example lies in the use of oral and topical probiotics, intended to limit the growth of pathogenic microorganisms while enhancing commensal ones (Krutmann, 2009; Ouwehand et al, 2003). Clinically, controlling the microbial community structure of the skin has the potential to decrease the rejection of viable skin grafts between individuals, as well as between different body locations within the same individual.
Identifying specific microbial community structure patterns of the human skin microbiota associated with disease will identify new potential intervention measures for improving health. It is anticipated that exploration of this new and different approach to human health will provide insights into disease etiology, management, and prevention.
4.3. Summary
Given the recent interest and technological advances in characterizing the human skin microbiota, it is important to learn whether certain diversity patterns or species composition of human microbiota are predictive or diagnostic of disease. A conceptual framework for understanding the interactions between skin microbiota, the human host and the environment is presented here in order to organize what host, dispersal, behavior, and environmental factors, or combination thereof, have the potential to drive the variability of the microbial community structure, thereby altering the skin microbiota diversity in such a way to cause disease.
Highlights.
We review the skin microbiota and its related methodological issues.
Host factors, behavior, environment, and transmission influence microbial community structure.
Human microbial communities impact health and disease.
We present a conceptual framework to explain the mediation of skin microbiota.
Acknowledgments
Role of the Funding Source: Support for MR was provided by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health funded Interdisciplinary Program in Infectious Diseases Training Grant (T32 AI049816).
Footnotes
Conflicts of Interest: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Deborah Goldberg, Email: degold@umich.edu.
Allison Aiello, Email: aielloa@umich.edu.
Elaine Larson, Email: ell23@columbia.edu.
Betsy Foxman, Email: bfoxman@umich.edu.
References
- 1.Aiello AE, Cimiotti J, Della-Latta P, Larson EL. A comparison of the bacteria found on the hands of ‘homemakers’ and neonatal intensive care unit nurses. J Hosp Infect. 2003;54(4):310–315. doi: 10.1016/s0195-6701(03)00146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiello AE, Coulborn RM, Perez V, Larson EL. Effect of hand hygiene on infectious disease risk in the community setting: a meta-analysis. Am J Public Health. 2008;98(8):1372–81. doi: 10.2105/AJPH.2007.124610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allegranzi B, Pittet D. Role of hand hygiene in healthcare-associated infection prevention. Hosp Infect. 2009;73:305–315. doi: 10.1016/j.jhin.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Bek-Thomsen M, Lomholt HB, Kilian M. Acne is Not Associated with Yet-Uncultured Bacteria. J Clin Microbiol. 2008;46(10):3355–3360. doi: 10.1128/JCM.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107(44):18933–8. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borges LFA, Silva BL, Filho PPG. Hand Washing: Changes in the skin flora. Am J Infect Control. 2007;35:417–20. doi: 10.1016/j.ajic.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Bowcock AM, Woodson WOCM. The genetics of psoriasis, psoriatic arthritis, and atopic dermatitis. Hum Mol Genet. 2004;13(1):R43–R55. doi: 10.1093/hmg/ddh094. [DOI] [PubMed] [Google Scholar]
- 8.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. Decreased Diversity of the Fecal Microbiome in Recurrent Clostridium difficile–Associated Diarrhea. J Infect Dis. 2008;197:435–8. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 9.Chiller K, Selkin BA, Murakawa GJ. Skin Microflora and Bacterial Infections of the Skin. J Invest Dermatol Symp Proc. 2001;6:170–174. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 10.Cogen AJ, Nizet V, Gallo RL. Skin microbiota: a source of disease or defence? Brit J Dermatol. 2007;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correa MC, Memije EV, Vargas-Alarcón G, Guzmán RA, Rosetti F, Acuña-Alonzo V, Martínez-Rodríguez N, Granados J. HLA-DR association with the genetic susceptibility to develop ashy dermatosis in Mexican Mestizo patients. J Am Acad Dermatol. 2007;56(4):617–20. doi: 10.1016/j.jaad.2006.08.062. [DOI] [PubMed] [Google Scholar]
- 12.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies CE, Wilson MJ, Hill KE, Stephens P, Hill M, Harding KG, Thomas DW. Use of molecular techniques to study microbial diversity in the skin: Chronic wounds reevaluated. Wound Rep Reg. 2001;9:332–340. doi: 10.1046/j.1524-475x.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 14.Dekio I, Hayashi H, Sakamoto M, Kitahara M, Nishikawa T, Suematsu M, Benno Y. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J Med Microbiol. 2005;54:1231–1238. doi: 10.1099/jmm.0.46075-0. [DOI] [PubMed] [Google Scholar]
- 15.Dekio I, Sakamoto M, Hayashi H, Amagai M, Suematsu M, Benno Y. Characterization of skin microbiota in patients with atopic dermatitis and in normal subjects using 16S rRNA gene-based comprehensive analysis. J Med Microbiol. 2007;56:1675–1683. doi: 10.1099/jmm.0.47268-0. [DOI] [PubMed] [Google Scholar]
- 16.Delwart EL. Viral metagenomics. Rev Med Virol. 2007;17:115–131. doi: 10.1002/rmv.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006;21(9):517–523. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Diez Roux AV, Aiello AE. Multilevel analysis of infectious diseases. J Infect Dis. 2005;191(Suppl 1):S25–33. doi: 10.1086/425288. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dotterud LK, Wilsgaard T, Vorland LH, Falk ES. The effect of UVB radiation on skin microbiota in patients with atopic dermatitis and healthy controls. Int J Circumpolar Health. 2008;67(2-3):254–260. doi: 10.3402/ijch.v67i2-3.18282. [DOI] [PubMed] [Google Scholar]
- 21.Evans CA. Persistent individual differences in the bacterial flora of the skin of the forehead: Numbers of Propionibacteria. J Invest Dermatol. 1975;64:42–46. doi: 10.1111/1523-1747.ep12540897. [DOI] [PubMed] [Google Scholar]
- 22.Evans CA, Crook JR, Strom MS. The bacterial flora of the forehead and back of Alaskan native villagers in summer and in winter. J Invest Dermatol. 1984;82(3):294–7. doi: 10.1111/1523-1747.ep12260391. [DOI] [PubMed] [Google Scholar]
- 23.Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci USA. 2009;106(38):16393–16399. doi: 10.1073/pnas.0908446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández L, Delgado S, Herrero H, Maldonado A, Rodríguez JM. The bacteriocin nisin, an effective agent for the treatment of staphylococcal mastitis during lactation. J Hum Lact. 2008;24(3):311–6. doi: 10.1177/0890334408317435. [DOI] [PubMed] [Google Scholar]
- 25.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105(46):17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foxman B, Goldberg D, Murdock C, Xi C, Gilsdorf JR. Conceptualizing Human Microbiota: From Multicelled Organ to Ecological Community. Interdisciplinary Perspectives Infect Dis. 2008;2008:1–5. doi: 10.1155/2008/613979. Article ID 613979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fredricks DN. Microbial Ecology of Human Skin in Health and Disease. JID Symp Proc. 2001;6(3):167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- 28.Gao Z, Tseng C, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. PNAS. 2007;104(8):2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Z, Tseng C, Strober BE, Pei Z, Blaser MJ. Substantial Alterations of the Cutaneous Bacterial Biota in Psoriatic Lesions. PLoS ONE. 2008;3(7):e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, Wolfsberg TG, Turner ML, Segre JA. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. Topographical and Temporal Diversity of the Human Skin Microbiome. Science. 2009;324:1190. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamady M, Knight R. Microbial Community Profiling for Human Microbiome Projects: Tools, Techniques, and Challenges. Genome Res. 2009;19(7):1141–52. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hrncir T, Stepankova R, Kozakova H, Hudcovic T, Tlaskalova-Hogenova H. Gut microbiota and lipopolysaccharide content of the diet influence development of regulatory T cells: studies in germ-free mice. BMC Immunol. 2008;9(65):1–11. doi: 10.1186/1471-2172-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465(7296):346–9. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 35.Jha AK, Gurung D. Seasonal variation of skin diseases in Nepal: A hospital-based annual study of out-patient visits. Nepal Med Coll J. 2006;8(4):266–8. [PubMed] [Google Scholar]
- 36.Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296(5570):1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 37.Kagan LJ, Aiello AE, Larson E. The role of the home environment in the transmission of infectious diseases. J Community Health. 2002;27(4):247–267. doi: 10.1023/A:1016378226861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kampf G, Kramer A. Epidemiologic Background of Hand Hygiene and Evaluation of the Most Important Agents for Scrubs and Rubs. Clin Microbiol Rev. 2004;17(4):863–893. doi: 10.1128/CMR.17.4.863-893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klaenhammer TR. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12(1-3):39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 40.Krutmann J. Pre- and probiotics for human skin. J Dermatol Sci. 2009;54:1–5. doi: 10.1016/j.jdermsci.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, Wu ZR, Hooper LV, Schmidt RR, von Aulock S, Radek KA, Huang CM, Ryan AF, Gallo RL. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15(12):1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larson EL, Cronquist AB, Whittier S, Lai L, Lyle CT, Della Latta P. Differences in skin flora between inpatients and chronically ill outpatients. Heart Lung. 2000;29(4):298–305. doi: 10.1067/mhl.2000.108324. [DOI] [PubMed] [Google Scholar]
- 43.Larson E. Hygiene of the skin: When is clean too clean? Emerg Infect Dis. 2001;7(2):225–230. doi: 10.3201/eid0702.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larson EL, Gomez-Duarte C, Lee LV, Della-Latta P, Kain DJ, Keswick BH. Microbial flora of hands of homemakers. Am J Infect Control. 2002;31(2):72–9. doi: 10.1067/mic.2003.33. [DOI] [PubMed] [Google Scholar]
- 45.Larson EL, Lin SX, Gomez-Pichardo C, Della-Latta P. Effect of antibacterial home cleaning and handwashing products on infectious disease symptoms: A randomized, double-blind trial. Ann Intern Med. 2004;140(5):321–329. doi: 10.7326/0003-4819-140-5-200403020-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of Engagement: Interspecies Interactions that Regulate Microbial Communities. Ann Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 47.Lo CW, Lai YK, Liu YT, Gallo RL, Huang CM. Staphylococcus aureus Hijacks a Skin Commensal to Intensify Its Virulence: Immunization Targeting β-Hemolysin and CAMP Factor. J Invest Dermatol. 2010 doi: 10.1038/jid.2010.319. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luby SP, Agboatwalla M, Feikin DR, Painter J, Billhimer WMS, Altaf A, Hoekstra RM. Effect of handwashing on child health: a randomised controlled trial. Lancet. 2005;366:225–33. doi: 10.1016/S0140-6736(05)66912-7. [DOI] [PubMed] [Google Scholar]
- 49.Mai V, Draganov PV. Recent advances and remaining gaps in our knowledge of associations between gut microbiota and human health. World J Gastroenterol. 2009;15(1):81–85. doi: 10.3748/wjg.15.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maier RM, Palmer MW, Andersen GL, Halonen MJ, Josephson KC, Maier RS, Martinez FD, Neilson JW, Stern DA, Vercelli D, Wright AL. The Bacterial Community in Household Dust: Environmental Determinants and Impact on Childhood Asthma. Appl Environ Microbiol. 2010;76(8):2663–2667. doi: 10.1128/AEM.01665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marris E. Staving off ecological disaster in lungs. 2009 http://www.nature.com/news/2009/090807/full/news.2009.808.html.
- 52.Martin-Visscher LA, van Belkum MJ, Garneau-Tsodikova S, Whittal RM, Zheng J, McMullen LM, Vederas JC. Isolation and characterization of carnocyclin a, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl Environ Microbiol. 2008;74(15):4756–63. doi: 10.1128/AEM.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marshall BM, Ochieng DJ, Levy SB. Commensals: Underappreciated Reservoir of Antibiotic Resistance. Microbe. 2008;4(5):231–238. [Google Scholar]
- 54.Masenga J, Garbe C, Wagner J, Orfanos CE. Staphylococcus aureus in atopic dermatitis and in nonatopic dermatitis. Int J Dermatol. 1990;29(8):579–82. doi: 10.1111/j.1365-4362.1990.tb03473.x. [DOI] [PubMed] [Google Scholar]
- 55.McBride ME, Duncan WC, Knowx JM. Cutaneous microflora of patients with repeated skin infections. J Cutaneous Pathol. 1977;4:14–22. doi: 10.1111/j.1600-0560.1977.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 56.McGuire AL, Colgrove J, Whitney SN, Diaz CM, Bustillos D, Versalovic J. Ethical, legal, and social considerations in conducting the Human Microbiome Project. Genome Res. 2008;18:1861–1864. doi: 10.1101/gr.081653.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakabayashi A, Sei Y, Guillot J. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med Mycol. 2000;38:337–41. doi: 10.1080/mmy.38.5.337.341. [DOI] [PubMed] [Google Scholar]
- 58.Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. Diversity of Human Vaginal Bacterial Communities and Associations with Clinically Defined Bacterial Vaginosis. Appl Environ Microbiol. 2008;74(15):4898–4909. doi: 10.1128/AEM.02884-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogunseitan O. Microbial Diversity. Blackwell Publishing; Malden, MA: 2005. [Google Scholar]
- 60.Oh S, Kim SH, Ko Y, Sim JH, Kim KS, Lee SH, Park S, Kim YJ. Effect of bacteriocin produced by Lactococcus sp HY 449 on skin-inflammatory bacteria. Food Chem Toxicol. 2006;44:1184–1190. doi: 10.1016/j.fct.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 61.Ordovas JM, Mooser V. Metagenomics: The role of the microbiome in cardiovascular diseases. Curr Opin Lipidol. 2006;17:157–161. doi: 10.1097/01.mol.0000217897.75068.ba. [DOI] [PubMed] [Google Scholar]
- 62.Othman M, Aguero R, Lin HC. Alterations in intestinal microbial flora and human disease. Curr Opin Gastroenterol. 2008;24:11–16. doi: 10.1097/MOG.0b013e3282f2b0d7. [DOI] [PubMed] [Google Scholar]
- 63.Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scandinavian J Gastroenterol. 2009;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 64.Otto M, Echner H, Voelter W, Götz F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69(3):1957–60. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ouwehand AC, Batsman A, Salminen S. Probiotics for the skin: a new area of potential application? Lett Appl Microbiol. 2003;36:327–331. doi: 10.1046/j.1472-765x.2003.01319.x. [DOI] [PubMed] [Google Scholar]
- 66.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular Analysis of Fungal Microbiota in Samples from Healthy Human Skin and Psoriatic Lesions. J Clin Microbiol. 2006;44(8):2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pivarcsi A, Nagy I, Kemeny L. Innate Immunity in the Skin: How Keratinocytes Fight Against Pathogens. Curr Immunol Rev. 2005;1:29–42. [Google Scholar]
- 69.Price LB, Liu CM, Johnson KE, Aziz M, Lau MK, Bowers J, Ravel J, Keim PS, Serwadda D, Wawer MJ, Gray RH. The effects of circumcision on the penis microbiome. PLoS One. 2010;5(1):1–12. doi: 10.1371/journal.pone.0008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rintala H, Pitkäranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 2008;8(56):1–13. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romero-Steiner S, Witek T, Balish E. Adherence of skin bacteria to human epithelial cells. J Clin Microbiol. 1990;28(1):27–31. doi: 10.1128/jcm.28.1.27-31.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roth RR, James WD. Microbial Ecology of the Skin. Ann Rev Microbiol. 1988;42:441–61. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- 73.Sawa N, Zendo T, Kiyofuji J, Fujita K, Himeno K, Nakayama J, Sonomoto K. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp strain QU 12. Appl Environ Microbiol. 2009;75(6):1552–8. doi: 10.1128/AEM.02299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scharschmidt TC, List K, Grice EA, Szabo R, NISC Comparative Sequencing Program. Renaud G, Lee CC, Wolfsberg TG, Bugge TH, Segre JA. Matriptase-deficient mice exhibit ichthyotic skin with a selective shift in skin microbiota. J Invest Dermatol. 2009;129(10):2435–42. doi: 10.1038/jid.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Selwyn S. Natural antibiosis among skin bacteria as a primary defence against infection. Brit J Dermatol. 1975;93:487–493. doi: 10.1111/j.1365-2133.1975.tb02241.x. [DOI] [PubMed] [Google Scholar]
- 76.Singh S, Kaye S, Gore ME, McClure MO, Bunker CB. The role of human endogenous retroviruses in melanoma. Brit J Dermatol. 2009;161:1225–1231. doi: 10.1111/j.1365-2133.2009.09415.x. [DOI] [PubMed] [Google Scholar]
- 77.Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16(3):107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 79.Sultana B, Cimiotti J, Aiello AE, Sloan D, Larson E. Effects of age and race on skin condition and bacterial counts on hands of neonatal ICU nurses. Heart Lung. 2003;32(4):283–9. doi: 10.1016/s0147-9563(03)00095-5. [DOI] [PubMed] [Google Scholar]
- 80.Theron J, Cloete TE. Molecular Techniques for Determining Microbial Diversity and Community Structure in Natural Environments. Crit Rev Microbiol. 2000;26(1):37–57. doi: 10.1080/10408410091154174. [DOI] [PubMed] [Google Scholar]
- 81.Tiwari SK, Srivastava S. Purification and characterization of plantaricin LR14: a novel bacteriocin produced by Lactobacillus plantarum LR/14. Appl Microbiol Biotechnol. 2008;79(5):759–67. doi: 10.1007/s00253-008-1482-6. [DOI] [PubMed] [Google Scholar]
- 82.Tlaskalová-Hogenová H, Štepánková R, Hudcovic T, Tucková L, Cukrowska B, Lodinová-Žádnıová R, Kozáková H, Rossmanna P, Bártová J, Sokol D, Funda DP, Borovská D, Reháková Z, Šinkora J, Hofman J, Drastich P, Kokešová A. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93:97–108. doi: 10.1016/j.imlet.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 83.van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, Wertheim HFL, Verbrugh HA. Co-evolutionary aspects of human colonization and infection by Staphylococcus aureus. Infect Genet Evol. 2009;9:32–47. doi: 10.1016/j.meegid.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 84.Wanke I, Steffen H, Christ C, Krismer B, Götz F, Peschel A, Schaller M, Schittek B. Skin Commensals Amplify the Innate Immune Response to Pathogens by Activation of Distinct Signaling Pathways. J Invest Dermatol. 2010 doi: 10.1038/jid.2010.328. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 85.Wilson M. Microbial Inhabitants of Humans: Their Ecology and Role in Health and Disease. first. Cambridge University Press; Cambridge: 2005. [Google Scholar]
- 86.Wilson M. Bacteriology of Humans: an ecological perspective. first. Blackwell Publishing; Oxford: 2008. [Google Scholar]
- 87.Wright P, Terry CS. Antagonism within populations of micro-organisms from normal human skin. J Med Microbiol. 1981;14:271–278. doi: 10.1099/00222615-14-3-271. [DOI] [PubMed] [Google Scholar]
- 88.Young VB, Britton RA, Schmidt TM. The human microbiome and infectious diseases: Beyond Koch. Interdisciplinary Perspectives on Infectious Diseases. 2008;2008:1–2. doi: 10.1155/2008/296873. Article ID 296873. [DOI] [PMC free article] [PubMed] [Google Scholar]


