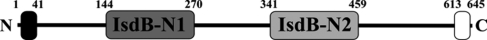Abstract
Iron is an essential requirement for life for nearly all organisms. The human pathogen Staphylococcus aureus is able to acquire iron from the heme cofactor of hemoglobin (Hb) released from lysed erythrocytes. IsdB, the predominant Hb receptor of S. aureus, is a cell wall-anchored protein that is composed of two NEAT domains. The N-terminal NEAT domain (IsdB-N1) binds Hb, and the C-terminal NEAT domain (IsdB-N2) relays heme to IsdA for transport into the cell. Here we present the 1.45 Å resolution X-ray crystal structure of the IsdB-N2–heme complex. While the structure largely conforms to the eight-strand β-sandwich fold seen in other NEAT domains such as IsdA-N and uses a conserved Tyr residue to coordinate heme-iron, a Met residue is also involved in iron coordination, resulting in a novel Tyr-Met hexacoordinate heme-iron state. The kinetics of the transfer of heme from IsdB-N2 to IsdA-N can be modeled as a two-step process. The rate of transfer of heme between the isolated NEAT domains (82 s–1) was found to be similar to that measured for the full-length proteins. Replacing the iron coordinating Met with Leu did not abrogate high-affinity heme binding but did reduce the heme transfer rate constant by more than half. This unusual Met-Tyr heme coordination may also bestow properties on IsdB that help it to bind heme in different oxidation states or extract heme from hemoglobin.
The Gram-positive bacterium Staphylococcus aureus is a common member of the human flora, and colonization with S. aureus primarily results in a commensal relationship with the host.1 Nevertheless, S. aureus is one of the main agents of nosocomial infections and can cause a range of disease, from mild (skin infections such as boils and folliculitis) to severe, life-threatening bloodstream infections.2S. aureus possesses a large number of virulence factors that contribute to its ability to cause infection, ranging from secreted toxins and adhesins to strategies for avoiding or subverting the innate and humoral immune responses, as well as the well-documented emergence of antibiotic resistance among increasing numbers of isolates.1,3
In addition to challenges from the innate and acquired immune systems, invading S. aureus must also contend with the nutritional immunity of the human body. Iron is an absolute requirement for diverse cellular processes in the human body, but free iron can destructively react with biological macromolecules; as a result, iron is tightly sequestered within the body.4 During infection, iron sequestration is significantly upregulated through several mechanisms (including enhanced expression of high-affinity iron chelators such as transferrin and lactoferrin) that render biological iron virtually inaccessible to invading pathogens, preventing them from usurping host iron to grow and infect.5 Successful human pathogens have therefore developed mechanisms for accessing sequestered host iron. The most abundant form of iron in the body is in complex with protoporphyrin IX, also known as heme, the majority of which is bound in the host oxygen-carrier molecule hemoglobin (Hb).4S. aureus exhibits a preference for heme as an iron source during infection6 and encodes a heme uptake system called the iron-responsive surface determinant (Isd) system, which is able to exploit the heme and hemoprotein resources of the host.7
The Isd system comprises five transcriptional units, each under the control of the Fur repressor, whose repression is relieved under iron-limiting conditions (for a review, see ref (8)). The system consists of: four peptidoglycan-anchored surface proteins that can all bind extracellular heme (IsdABCH), a membrane ABC transporter with associated lipoprotein (IsdEF), and two intracellular heme-degrading enzymes (IsdGI). The surface proteins of the Isd system are C-terminally covalently anchored to the peptidoglycan by sortase A (in the case of IsdABH) or sortase B (in the case of IsdC). Furthermore, the surface proteins each contain one to three copies of a conserved protein fold known as a NEAT (for NEAr iron transporter) domain, a fold of ∼125 residues that was identified bioinformatically as being in the genomic neighborhood of putative iron uptake systems.9 Although the degree of sequence identity between NEAT domains can vary wildly, crystal structures of Isd NEAT domains have revealed a common eight-strand β-sandwich fold.10−12 The NEAT domains of the surface Isd proteins have been further demonstrated to mediate either heme binding10−16 or hemoprotein binding.14,17,18
IsdB contains two NEAT domains9 with 12% pairwise sequence identity and acts as the primary Hb receptor for the cell.19 Deletion of isdB virtually abolishes Hb binding by the S. aureus cell and significantly impacts disease in a mouse infection model.19 IsdB is also highly immunogenic during infection and is the basis for a monovalent vaccine currently undergoing clinical trials.20 IsdB extracts the heme from Hb at the cell surface for the transfer to IsdA or IsdC, which then transfers it to the membrane transporter for internalization.15,21 Recent evidence strongly implies that each NEAT domain of IsdB has a different binding specificity; note that NEAT domain numbering proceeds from the N-terminus to the C-terminus (Figure 1). IsdB-N1 binds Hb but not heme.22 Conversely, IsdB-N2 binds heme15,16 but not Hb,22 and its sequence contains a conserved Tyr residue that is known to coordinate the heme-iron in all other heme binding NEAT domains.10−12 IsdB-N2 alone is able to transfer heme to IsdA-N1 and IsdC-N1;15 however, full-length IsdB transfers heme to full-length IsdA at 8 times the rate for full-length IsdC,21 and protease digestion suggests that IsdB may be physically closer to IsdA than IsdC on the cellular surface,7 suggesting the physiological route of heme transfer is likely from IsdB-N2 to IsdC-N1 via IsdA-N1.
Figure 1.
Schematic of IsdB sequence features. Amino acids are numbered according to the S. aureus N315 IsdB sequence (NP_374246). The black bar near the N-terminus signifies the signal sequence, which directs the protein to be secreted onto the surface. The white bar near the C-terminus signifies the LPQTG sortase signal, which directs the protein to be covalently anchored to the peptidoglycan by sortase A. The two NEAT domains are shown in different shades of gray to indicate their dissimilar pairwise sequence identity (12%). The mature protein on the staphylococcal surface encompasses Ala41–Thr613.
We sought to characterize heme binding by IsdB-N2 as a first step in understanding Hb reception and heme transfer by the Isd system. We determined the 1.45 Å resolution crystal structure of heme-bound IsdB-N2 and discovered a new mode of heme-iron coordination, with protein ligands provided by both a Met and a Tyr. We further showed that iron coordination by the Met residue is dispensable for high-affinity heme binding but has a role in facilitating the transfer of heme to IsdA-N1.
Experimental Procedures
Cloning of NEAT Domains and Variants
The coding region of the second, heme binding NEAT domain (IsdB-N2, Lys341–Thr459) was amplified from S. aureus N315 chromosomal DNA and cloned into the pET28a(+) expression vector for expression with an N-terminal His6 tag and thrombin cleavage site. Site-directed variants were created by subcloning from the IsdB-N2 clone, using a modified whole plasmid polymerase chain reaction method.23 All clones were verified by sequencing (Agencourt, Beverly, MA).
The NEAT domain coding region of IsdA (IsdA-N1, Ser62–Ala184) was subcloned from the previous construct in pET28a, complete with the N-terminal His6 tag and ribosome binding site,10 into pBAD-1824 using the restriction digestion enzymes XbaI and HindIII as previously described.13
Protein Expression and Purification
Recombinant IsdB-N2 was overexpressed in Escherichia coli BL21(DE3) cells. A 2 L bacterial culture was grown from 2 mL of overnight culture in Luria-Bertani (LB) broth supplemented with 25 μg/mL kanamycin at 30 °C to an OD600 of 0.7–0.9, then induced with 0.5 mM isopropyl β-d-thiogalactopyranoside, and grown for an additional 18 h at 25 °C. Cells were pelleted by centrifugation, resuspended in 20 mL of 20 mM sodium phosphate (pH 7.4) and 500 mM NaCl, and then lysed at 4 °C using an EmulsiFlex-C5 homogenizer (Avestin, Ottawa, ON). Insoluble material was removed by centrifugation; the soluble lysate contained a mixture of apo- and holo-His6-IsdB-N2, and apoprotein could be separated at 4 °C using a HisTrap nickel affinity column (GE Healthcare) by elution with an imidazole gradient. The apoprotein was dialyzed against 50 mM Tris (pH 8.0) and 100 mM NaCl then cleaved with thrombin at a 1:500 ratio by weight of His6-IsdB-N2 to remove the His6 tag. IsdB-N2 was then dialyzed against 50 mM MOPS (pH 7.0) for cation exchange chromatography using a Source 15S column (GE Healthcare), and purified protein was obtained by elution with a NaCl gradient. The resulting pure (>95% by sodium dodecyl sulfate–polyacrylamide gel electrophoresis) apoprotein was dialyzed against either 20 mM Tris (pH 8.0) for crystallization or 50 mM Tris (pH 8.0) and 100 mM NaCl for spectroscopic and kinetic studies.
Selenomethionine-labeled (Se-Met) IsdB-N2 was prepared by the method previously described by Van Duyne et al.25 and purified as described for native protein.
Heme Reconstitution
Purified apoprotein was incubated for 1 h at 4 °C with 1.5 molar equiv of hemin dissolved in 0.1 M NaOH and diluted in 0.1 M phosphate buffer (pH 7.4). Excess hemin was removed by centrifugation, and nonspecifically bound hemin was removed by gel filtration chromatography on a Sephadex G-50 column (1 cm × 6 cm). The concentration of the holoprotein was then determined by the pyridine hemochrome assay using an ε418 extinction coefficient of 191.5 mM–1 cm–1 as previously described.26
Crystal Structure Determination
Native holo-IsdB-N2 and Se-Met IsdB-N2 crystals were grown by hanging drop vapor diffusion at room temperature at a 1:1 ratio of protein to well solution [composed of 0.1 M Tris (pH 8.0), 0.1 M MgCl2, and 25% polyethylene glycol 3350]. Crystals were briefly washed in mother liquor and flash-frozen in liquid nitrogen. Data were collected at the Stanford Synchrotron Radiation Lightsource on beamline 9-2. Data were processed and scaled using HKL2000.27 Crystals grew in space group P212121 with four molecules in the asymmetric unit.
Se-Met crystals were generated initially for anomalous phasing, but in the interim, a suitable model was deposited in the Protein Data Bank (PDB) (IsdH-N3, 60% identical; PDB entry 2E7D) and molecular replacement with a single chain (with no heme) from that structure using MolRep28 from the CCP4 program suite29 yielded interpretable phases. Se-Met IsdB-N2 was built using ARPWarp,30 and manual building was completed using Coot.31 A Ramachandran plot revealed that 92.5% of residues were in the most favored regions with the remaining 7.5% in additional allowed regions. Subsequently, native holo-IsdB-N2 crystals were generated, and MolRep was again used for phasing by molecular replacement with the Se-Met structure as the search model. The structure was modified using Coot31 and refined with Refmac5.32 Multiple conformations of side chains were modeled by visual examination of Fo – Fc maps. Waters were added using the ARP/Waters30 function in Refmac5, and model quality parameters were assessed using Procheck.33 A Ramachandran plot revealed that 94.3% of residues were in the most favored regions with the remaining 5.7% in additional allowed regions. Figures are of the native structure and were generated using PyMOL.34 Data collection and refinement statistics for both the native and Se-Met IsdB-N2 structures are listed in Table 1.
Table 1. X-ray Data Collection and Refinement Statistics.
| native | Se-Met | |
|---|---|---|
| Data Collectiona | ||
| space group | P212121 | P212121 |
| unit cell dimensions a, b, c (Å) | 57.5, 82.6, 116.9 | 52.8, 79.4, 113.9 |
| resolution range (Å) | 50.0–1.45(1.50–1.45) | 50.0–1.70(1.76–1.70) |
| Rmerge | 0.05 (0.37) | 0.05 (0.31) |
| I/σI | 23.0 (3.9) | 24.2 (3.8) |
| completeness (%) | 99.4 (95.2) | 99.3 (95.5) |
| redundancy | 7.1 (5.7) | 4.6 (3.7) |
| no. of unique reflections | 97666 | 53106 |
| Wilson B (Å2) | 18.5 | 23.6 |
| Refinement | ||
| Rwork/Rfree | 0.178/0.217 | 0.193/0.232 |
| no. of atoms | 4993 | 4793 |
| protein/heme | 4364 | 4033 |
| ions | 6 | 5 |
| water | 623 | 755 |
| B factor (Å2) | ||
| protein | 19.5 | 26.7 |
| heme | 15.0 | 22.8 |
| ions | 17.1 | 23.5 |
| water | 28.3 | 42.0 |
| root-mean-square deviation | ||
| bond lengths (Å) | 0.012 | 0.013 |
| bond angles (deg) | 1.38 | 1.48 |
Values in parentheses represent those for the highest-resolution shell.
Determination of the IsdB-N2 Heme Binding Stoichiometry
Heme binding was tracked by difference absorption spectroscopy in the Soret region (around 400 nm) at room temperature. Aliquots of hemin (1.3 μL) solubilized in 0.1 M NaOH and diluted in 50 mM Tris (pH 8.0) and 100 mM NaCl were added to 1 mL of a 5 μM solution of apo-IsdB-N2 or a reference cuvette containing buffer alone. Spectra were recorded 10 min after addition of heme, and titrations covered a heme concentration range from 0.5 to 12 μM. Saturation was defined as a plateau in the absorption difference between the reference cuvette and the experimental cuvette. For reference, absorption spectra (250–650 nm) of purified reconstituted proteins were measured. All spectra were recorded using a Cary 50 Bio UV–visible spectrophotometer (Agilent Technologies, Mississauga, ON) with an optical path length of 1 cm in a quartz cell at room temperature.
Trp Fluorescence Quenching by Heme
Heme binding was monitored by fluorescence quenching of the tryptophan residue (Trp392) at the base of the heme pocket, based on the method described by Eakanunkul et al.35 Fluorescence emission spectra from 300 to 450 nm were recorded at 20 °C with excitation at 295 nm in 50 mM Tris (pH 8) and 100 mM NaCl with an apo-IsdB-N2 concentration of 1 μM in a 1 mL volume using a Cary Eclipse fluorescence spectrophotometer (Agilent Technologies). Heme (prepared as described above, in various dilutions) was added to the buffered protein solution in 0.5–1.5 μL aliquots and allowed to reach equilibration (which ranged from 6 to 12 min depending on the variant) before readings were taken. The titrations covered a heme concentration range of 0.1–15 μM. The dissociation constant (KD) was calculated from the decrease in the fluorescence intensity between 300 and 450 nm as a function of increasing heme concentration, using an equation for equilibrium binding that accounts for ligand depletion as described by Stein et al.36 All reactions were conducted in triplicate.
Determination of Rates of Transfer of Heme to Apomyoglobin
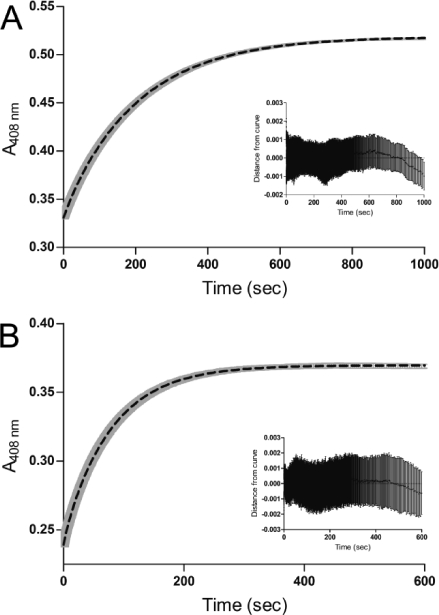
The rates of dissociation of heme from IsdB-N2 were measured by single-wavelength stopped-flow spectroscopy with apomyoglobin (apoMb) as a heme scavenger.37 ApoMb was prepared from myoglobin (Sigma-Aldrich, St. Louis, MO) as previously described.38 Heme dissociation reactions were conducted with 10 μM holo-IsdB-N2 (reconstituted as described above) in one syringe (final concentration of 5 μM) and 100 μM apomyoglobin (final concentration of 50 μM) in the second syringe, both in 50 mM Tris (pH 8) and 100 mM NaCl at room temperature in an SX.18MV stopped-flow reaction analyzer (Applied Photophysics, Leatherhead, U.K.). Reactions were monitored by recording the absorbance at 408 nm for 1000 s (wild-type; ∼7 half-lives) or 600 s (M362L variant; ∼11 half-lives); 1000 readings at logarithmic intervals were acquired using Pro-Data SX software, regardless of the time frame. The change in absorbance at 408 nm (maximal absorbance for holomyoglobin) was plotted versus time and fit by a double-exponential equation to yield the first-order rate constants for heme dissociation using GraphPad Prism version 5.02 for Windows (GraphPad Software, La Jolla, CA). All reactions were conducted in triplicate.
Determination of Rates of Transfer of Heme to IsdA-N1
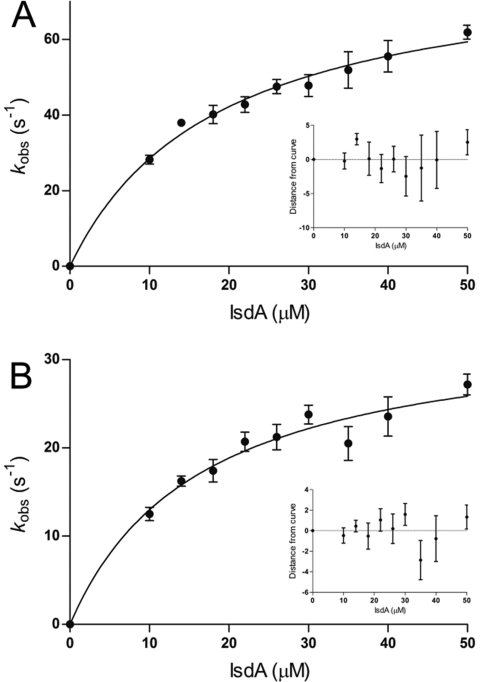
Differences in spectral characteristics between holo-IsdB-N2 and holo-IsdA-N1 were exploited to monitor the transfer of heme from holo-IsdB-N2 to apo-IsdA-N1 by stopped-flow spectroscopy, conducted using an SX.18MV stopped-flow reaction analyzer (Applied Photophysics) equipped with a photodiode array detector. The reactions were conducted with 4 μM holo-IsdB-N2 in one syringe (final concentration of 2 μM) and concentrations of apo-IsdA-N1 ranging from 20 to 100 μM (final concentrations of 10–50 μM) in the second syringe; a minimal 5-fold excess of the IsdA acceptor was used to attain pseudo-first-order conditions. One hundred spectra were recorded at logarithmic intervals from 0 to 10 s from approximately 300 to 650 nm, using Xscan (Applied Photophysics). The drive syringe chamber and optical cell were maintained at 25 °C by a circulating water bath. The change in absorbance over time at the wavelength of maximal change (418.9 nm for the wild type and 425.3 nm for the M362L variant) for a given concentration was fit by a single-exponential equation to determine the observed transfer rate (kobs). All reactions were conducted in quadruplicate.
Results
Crystal Structure of IsdB-N2
The holo structure of IsdB-N2 was determined to 1.45 Å resolution. The structure of Se-Met IsdB-N2 was also determined to 1.7 Å but was inappropriate for interpretation of heme binding due to the artifact introduced by the selenomethionine substitution, discussed in a later section. There were four molecules in the asymmetric unit that overlaid with an average root-mean-square deviation (rmsd) of 0.49 Å over all Cα atoms, with the main differences occurring at the N-termini. The final model consists of residues 341–452 for chain A, 341–458 for chains B and C, and 341–456 for chain D and includes an N-terminal Gly-Ser cloning artifact left over from the thrombin cleavage site.
IsdB-N2 adopts the characteristic eight-strand immunoglobulin-like β-sandwich fold observed in other NEAT domains whose structures are known (Figure 2A).10−12,14,39 In addition to short α-helices between β1 and β2 and between β3 and β4, there is a C-terminal α-helix of approximately 1.5 turns immediately following β8 that is not observed in other NEAT domain structures.
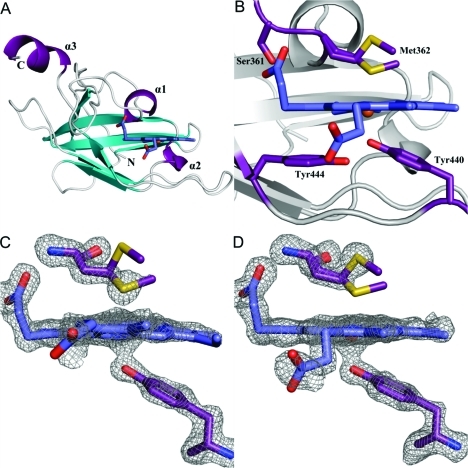
Figure 2.
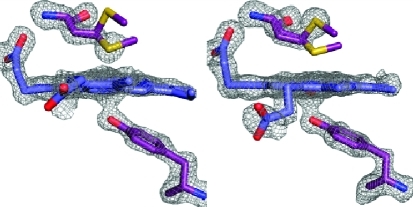
Crystal structure of the heme-bound C-terminal NEAT domain of IsdB. (A) Overall structure of holo-IsdB-N2 (chain A) viewed down the heme binding pocket. The backbone is shown as a cartoon with helices colored purple, β-strands cyan, and loops gray. Heme is shown protruding from the pocket as sticks, with carbon atoms colored dark blue and oxygen atoms red. One conformation of heme is shown for the sake of clarity. The N- and C-termini and helices are labeled. (B) Close-up of the heme pocket. Residues directly involved in binding the heme molecule (Tyr440, Met362, and Ser361) or indirectly involved in binding (Tyr444) are shown as sticks and labeled. Protein carbon, oxygen, and sulfur atoms are colored purple, red, and yellow, respectively. Heme carbon, oxygen, nitrogen, and iron atoms are colored blue, red, dark blue, and dark red, respectively. (C and D) Fo – Fc omit maps (contoured at 3σ) for Tyr440, Met362, and heme of chains A and B.
Structure of the Heme Binding Pocket
Heme is bound in a highly hydrophobic pocket and is modeled at equal occupancy in two orientations when rotated by 180° along the Cα–Cγ axis (Figure 2B). Approximately 330 Å2 (38%) of the heme surface area was found to be exposed to solvent as calculated using AreaIMol.29 Crystal packing interactions occurred across the face of the heme pocket, resulting in vinyl groups of heme molecules being ∼3.3 Å apart (Figure 1 of the Supporting Information). The heme propionates are extended out from the pocket, and one forms a hydrogen bond with the hydroxyl group of Ser361 and the backbone nitrogen of Met362. Tyr440, Tyr444, and Phe366 contribute to π-stacking with a buried heme pyrrole ring, whereas Val431, Val433, Val446, Val435, Tyr391, Trp392, and Met363 contribute to hydrophobic contacts.
The heme iron is coordinated by Tyr440, which forms a hydrogen bond with the phenolate of Tyr444, an interaction that is absolutely conserved among heme binding NEAT domains of known structure.10−12 The heme-iron exists in a mixture of hexacoordinate and pentacoordinate states, with the sulfur from Met362 occupying the sixth-coordinate position. The electron density is best modeled with Met362 in both coordinating and noncoordinating conformations in the structure, and the occupancy of the conformations is unequal and dependent upon the molecule examined. In chains A and D, inspection of Fo – Fc maps indicates that Met362 exists in a coordinating position at 75% occupancy (sulfur atom 2.5–2.6 Å from heme-iron) and 25% occupancy in a noncoordinating position (sulfur atom 4.9–5.0 Å from heme-iron) (Figure 2C). The opposite is true for chains B and C, where the sulfur atom models away from the pocket at 70% occupancy (4.8 Å from heme-iron) and the sulfur atom is directed toward the heme iron at 30% occupancy (2.6 Å from heme-iron) (Figure 2D). Furthermore, these conformations appear to be complementary to one another, such that the heme pocket containing a mainly coordinating Met362 (chain A or D) is tightly packed against a heme pocket containing a mainly noncoordinating Met362 (chain B or C). Lastly, the heme-iron is pulled out of the plane of the porphyrin ring and closer to Tyr440 in chains B and C (2.1–2.2 Å Fe–O bond length, 0.4–0.5 Å from planarity), where Met362 is mainly noncoordinating, whereas the iron is pulled away from Tyr440 and lies closer to the porphyrin ring plane in chains A and D (2.2–2.3 Å Fe–O bond length, less than 0.2 Å from planarity), where Met362 is mainly coordinating.
The variability in the Met362 coordination state was first observed in the 1.7 Å Se-Met structure, motivating the determination of the native structure. Although it is well-documented that substitution of a Met heme-iron ligand for Se-Met has little effect on heme iron coordination,40 we were concerned that the slightly larger van der Waals radius of selenium or an incomplete substitution of Se-Met for Met might have caused the mixture of coordination states observed in the structure.
Visible Absorption Spectra of Heme Pocket Variants
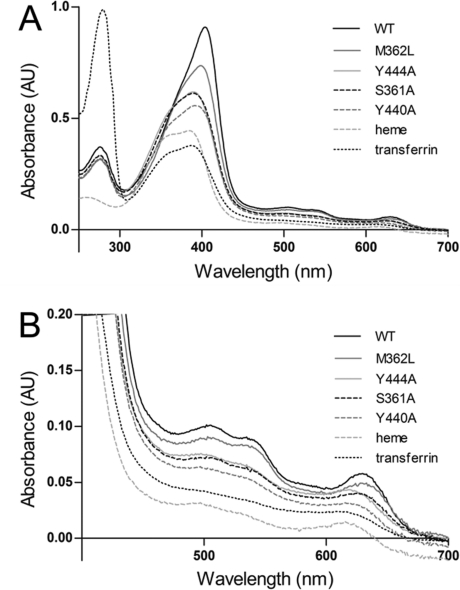
On the basis of the residues that were seen to directly participate in heme binding in the IsdB-N2 crystal structure, several variants of the wild-type (WT) protein were constructed using site-directed mutagenesis: Y440A, Y444A, S361A, and M362L. The visible absorption spectra of the variants reconstituted with equimolar hemin were compared to that of the WT protein, as the spectrum is indicative of the heme environment. Mutation of any one of these residues was found to significantly affect the shape, height, and maximum wavelength of the Soret peak, indicating a significant change in the environment experienced by the heme (Figure 3A). Furthermore, the spectra of the Y444A, Y440A, and S361A variants more closely resembled that of free heme than that of WT IsdB-N2, indicating that loss of Tyr444, Tyr440, or Ser361 resulted in severe heme binding disruption. The spectrum of M362L resembled most closely that of the wild-type hemoprotein rather than that of free heme, with a less pronounced shoulder at 380 nm and more pronounced α/β bands (Figure 3B). However, the Soret peak shifted from 404 nm for WT to 398 nm for M362L, and the Soret shape and height were also significantly different, indicating different heme environments.
Figure 3.
Visible spectra of 5 μM holo-IsdB-N2 WT and variants. Heme (5 μM) was added to apoprotein (5 μM) and allowed to reach equilibrium at room temperature. Spectra of heme (5 μM) alone in buffer and heme (5 μM) added to transferrin, a non-heme binding protein, are also shown for comparison. (A) Spectra of all proteins recorded between 250 and 700 nm. The absorbance at 280 nm is similar for all IsdB-N2 proteins (∼14 kDa), whereas that of transferrin (∼80 kDa) is significantly higher; heme absorbs much less at 280 nm. The spectrum in the Soret range (∼400 nm) is noticeably different for most variants, except S361A and Y444A, which are largely indistinguishable. (B) Close-up of the spectra between 400 and 700 nm revealing characteristic markers of the heme-iron environment. Spectra of WT and M362L are most similar to each other, whereas the spectra of Y440A, Y444A, and S361A resemble the spectrum of free heme.
Heme Titration of IsdB-N2 and Variants
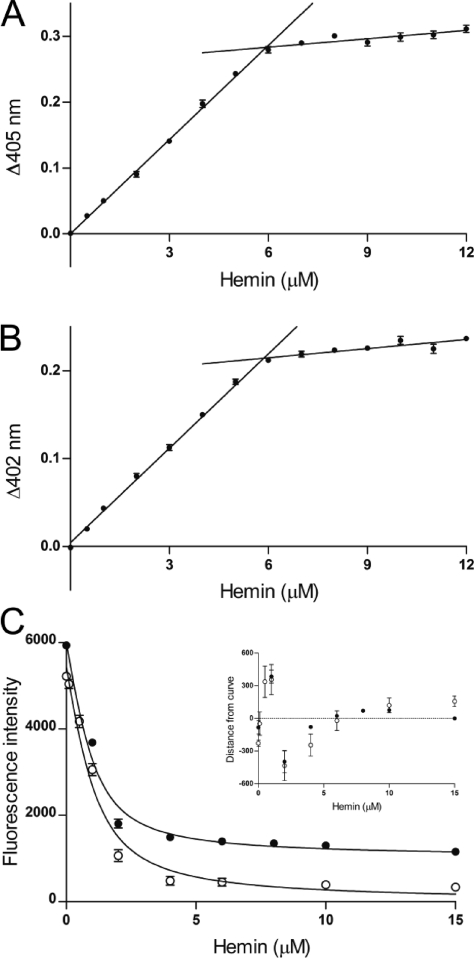
Although a single heme molecule is bound per monomer of IsdH-N3 in the crystal structure, investigators found that a monomer could bind up to four molecules of heme in solution.12 Though the crystal structure of IsdB-N2 also bound one heme per monomer, to confirm that 1:1 stoichiometry was recapitulated in solution apoprotein was titrated with increasing amounts of heme and difference spectra were generated by subtracting spectra from that of the buffer titrated with the same amounts of heme. WT IsdB-N2 and M362L (Figure 4A,B) both demonstrated a binding stoichiometry of approximately 1:1. However, Y444A, Y440A, and S361A difference spectra did not follow a single-site, specific binding curve and resembled that of a non-heme binding protein, transferrin, indicating aberrant or extremely weak heme binding (data not shown). Because of the apparent weak binding for these variants, they were deemed unsuitable for performing transfer assays.
Figure 4.
IsdB-N2 WT and M362L variant follow a similar heme binding curve. Titration of 5 μM apo-IsdB-N2 wild-type protein (A) or M362L (B) with heme demonstrates a binding stoichiometry of ∼1:1, as the increase in absorbance at respective Soret peaks when compared to heme alone plateaus at approximately 6 μM. (C) Titration of 1 μM apo-IsdB-N2 WT protein (●) or M362L variant (○) with 0.1–15 μM heme results in a decrease in Trp fluorescence with an increasing heme concentration in a one-site specific manner. A graph of the residuals for both curves is inset.
IsdB-N2 contains one Trp residue (W392), which resides at the base of the heme binding pocket. We were able to exploit this fact to develop a fluorescence-based measurement of heme binding as heme addition causes quenching of Trp fluorescence. We observed a concentration-dependent and saturable quenching of Trp fluorescence by both WT IsdB-N2 and the M362L variant (Figure 4C). At a concentration of 1 μM, IsdB-N2 WT and M362L variant had very similar KD values (0.38 ± 0.06 and 0.49 ± 0.09 μM, respectively) when fit to an equation accounting for ligand depletion; however, as we are working at a protein concentration close to the calculated KD (necessarily high for a reproducible fluorescence signal), these values can be considered only an upper bound, with the actual KD likely being lower. We can conclude that both the native form and M362L variant bind heme with high nanomolar or better affinity. By comparison, the variant proteins that exhibited aberrant or weak heme binding by visible spectroscopy required much greater (at least 5-fold higher) heme concentrations for maximal quenching, and curve fitting suggested dissociation constants at least 5-fold higher (data not shown).
Rate of Transfer of Heme to Apomyoglobin
The transfer of heme from the binding pockets of IsdB-N2 WT and M362L variant to apomyoglobin (apoMb) was followed by visible stopped-flow spectroscopy at 408 nm, the Soret maximum for holomyoglobin. ApoMb has an affinity for heme in the picomolar range,41 does not directly bind to IsdB,19 and has been used to characterize heme binding by Isd proteins previously.21 The rate of transfer of heme to apoMb is independent of the concentration of apoMb; therefore, the observed rate is assumed to be the rate of release of heme (off rate) from IsdB-N2.37 The transfer rate data are best fit by a double-exponential curve, as judged by the greater randomness of the residual plot than that for a single-exponential curve. The initial fast phase accounted for only 2.8% of the curve for WT and 3.5% for M362L and may represent the presence of a minor extraneous species in solution, such as improperly folded holo-IsdB-N2 or IsdB-N2 with nonspecifically bound surface heme. The slow phase, describing the vast majority of the absorption change, yielded an off rate of 4.8 × 10–3 s–1 for WT IsdB-N2 (Figure 5A), and the substitution of Met362 with Leu (Figure 5B) increased the off rate by 2.7-fold, to 1.3 × 10–2 s–1. A summary of kinetic and equilibrium binding characteristics can be found in Table 2.
Figure 5.
Transfer of heme from IsdB-N2 to apomyoglobin. There is a considerable change in the visible spectrum as heme is released from holo-IsdB-N2 and bound by apomyoglobin (apoMb). (A and B) The increase in absorbance at 408 nm is characteristic of holomyoglobin formation; a plot of the absorbance at 408 nm vs time fits a two-phase association function in which the rate of formation of holomyoglobin is equal to the off rate from IsdB-N2. The dotted black line is a double-exponential curve fit to the data, and the gray band represents the standard error of three replicates for WT and two replicates for M362L. A graph of the residuals is inset and displays a relatively random distribution: (A) IsdB-N2 WT and (B) IsdB-N2 M362L.
Table 2. Kinetic Parameters of Heme Binding and Transfer.
| off rate ± SE (×10–3 s–1) |
|||||
|---|---|---|---|---|---|
| KD ± SE (heme; μM) | kfast | kslow | ktransfer ± SE (s–1) | KDa ± SE (μM) | |
| IsdB-N2 WT | <0.38 ± 0.06 | 28 ± 3 | 4.8 ± 0.02 | 81.5 ± 5.7 | 18.6 ± 3.2 |
| IsdB-N2 M362L | <0.49 ± 0.09 | 130 ± 11 | 13 ± 0.04 | 34.4 ± 2.4 | 16.5 ± 3.5 |
Estimated dissociation constant for the protein complexed with IsdA-N1.
Rate of Transfer of Heme to IsdA-N1
Using stopped-flow spectroscopy, the observed rates (kobs) of transfer of heme from holo-IsdB-N2 to apo-IsdA-N1 were determined by the difference in the Soret region of their visible spectra at the wavelength of maximal change over ∼0.3 s (for WT) or ∼1.0 s (for M362L), equivalent to a minimum of 15 half-lives. The change in absorbance at 418.9 nm (WT) or 425.3 nm (M362L) was plotted versus time and fit by a single exponential to determine a value for the observed rate constant (kobs). The kobs for 2 μM IsdB-N2 was determined under pseudo-first-order conditions for concentrations of IsdA-N1 from 10 to 50 μM. The kobs values vary hyperbolically with respect to the concentration of IsdA-N1, suggesting a two-step transfer mechanism. A model for heme transfer has been proposed by Liu et al.,42 which states that the transfer of heme from the holo-NEAT domain to the apo-NEAT domain has two observable steps characterized by rapid formation of the protein–protein complex followed by rate-limiting heme transfer between the proteins. Using this model, the rate constant from wild-type holo-IsdB-N2 to apo-IsdA-N1 was found to be 82 ± 6 s–1 (Figure 6A). For IsdB-N2 M362L to IsdA-N1, the rate constant was found to decrease by more than 2-fold, to 34 ± 3 s–1 (Figure 6B). The calculated KD for the protein–protein complexes was similar for both WT and M362L transfer reactions (18 and 16 μM, respectively).
Figure 6.
Kinetics of transfer of heme from holo-IsdB-N2 to apo-IsdA-N1. The observed transfer rate (kobs) varies hyperbolically with IsdA concentration, characteristic of a two-step reaction. Each point represents the mean and standard error of four replicates of the 2 μM IsdB-N2 to IsdA-N1 heme transfer experiment. Observed transfer rates at each concentration of IsdA-N1 were determined by plotting the change in absorbance at a given wavelength (418.9 nm for WT and 425.3 nm for M362L) vs time. The residuals for the curves are shown in the insets. (A) Transfer from WT holo-IsdB-N2 to apo-IsdA-N1. (B) Transfer from holo-IsdB-N2 M362L to apo-IsdA-N1.
Discussion
Investigating the molecular basis for the uptake of heme by S. aureus is a crucial step toward understanding host–pathogen interactions. IsdB is the dominant Hb-binding protein of S. aureus.19 We have found that the heme binding NEAT domain (IsdB-N2) of IsdB adopts a fold similar to those observed in other NEAT domain structures.10−12,14,17 Additionally, heme-iron is coordinated by a conserved Tyr residue as predicted by multiple-sequence alignments10 and observed in other heme binding NEAT domains in S. aureus.10−12 However, a unique feature of IsdB-N2 is a distal heme-iron ligand, Met362. The equivalent position in IsdA-N1 is taken by a His, which is a common iron ligand but does not coordinate to the heme-iron in the crystal structure10 or in solution in the absence of a strong reductant.16 In the heme binding NEAT domains of IsdC (IsdC-N1) and IsdH (IsdH-N3), there are an Ile and a Val, respectively, neither of which has the capacity for heme-iron coordination.11,12 As well as being unprecedented in the Isd system of S. aureus, Tyr-Met heme-iron coordination has not been reported in the literature for any heme binding protein to date.
The protein Shp from Streptococcus pyogenes uses an unusual bis-methionyl heme-iron coordination as part of a heme uptake system with some components distantly related to the Isd system of S. aureus.39 The crystal structure of the NEAT-like domain of Shp, denoted Shp180, superposes on IsdB-N2 with a core rmsd of 2.5 Å over 94 Cα atoms using SSM Superposition.43 The average Fe–S bond length between the sulfur of the coordinating Met residues of Shp and the heme-iron was found to be 2.4 Å,39 shorter than the Fe–S bond length of 2.5–2.6 Å found here. By comparison, the Fe–S bond length seen in IsdE, which coordinates heme-iron using Met and His, is shorter at 2.3 Å.40 The Fe–O bond length (Tyr–heme-iron) observed in IsdB-N2, which ranges from 2.1 to 2.3 Å depending on the molecule, is typical of those seen in other NEAT domain structures: 2.1 Å for IsdA-N1,40 2.2 Å for IsdH-N3,12 and 2.0–2.1 Å for IsdC-N1.11
It is noteworthy that previous work using electronic and magnetic circular dichroism (MCD) spectroscopy did not identify Met as a heme-iron ligand in IsdB-N2, though Tyr was accurately predicted to be a ligand.15,16,44 The discrepancy may be explained by photoreduction of the heme-iron to Fe2+ in the crystal by the X-ray radiation, resulting in a preference for ligation by methionine; beamline photoreduction is a well-known issue when studying hemoproteins and can result in conformational changes as well as changes in coordination state.45 However, IsdA-N1 contains a potential distal heme-iron ligand (His83) that was not observed to coordinate heme-iron in the crystal structure.10 Moreover, it has been shown that reduction of the heme-iron in IsdA results in heme iron coordination solely by the distal His83.13,46 Given that Met362 clearly occupies both coordinating and noncoordinating conformations in the IsdB-N2 crystal structure, we suggest that it may do the same in solution, consistent with spectra indicating a predominantly five-coordinate heme-iron with the sixth position readily available to bind exogenous CN–.44
Aside from Tyr440 and Met362, two other residues form important interactions to secure heme: Ser361, which hydrogen bonds to one propionate, and Tyr444, which hydrogen bonds with Tyr440. All four residues were mutated separately to probe their involvement in stable heme binding by IsdB-N2, resulting in the Y440A, Y444A, M362L, and S361A variants. Of these four, only the M362L variant was proficient at heme binding: a gel filtration step intended to purify hemoprotein after heme reconstitution resulted in mainly apoprotein in the cases of the Y440A, Y444A, and S361A variants (data not shown). The apparent deficiency in heme binding of these variants was further characterized by examining their visible electronic spectra, which more closely resembled that of free heme than that of hemoprotein. The loss of high-affinity heme binding by the S361A variant suggests that the interaction with the heme propionate is crucial for the stability of the loop that includes Met362 and forms one side of the heme binding pocket. Similarly, Tyr444 is absolutely conserved in all NEAT domains of the Isd system, heme binding or not; the data suggest that it is a critical heme binding residue as well, although it does not directly interact with the heme. Tyr444 may be required to stabilize the fold of NEAT domains as well as to position Tyr440 for heme-iron coordination or to mediate the phenolate–iron bond.
To investigate the significance of Met362 with regard to potential functional roles in the heme pocket, we conducted a series of spectroscopic and kinetic characterizations of the WT IsdB-N2 protein in comparison with an M362L variant. We found that the variant retained at minimum low micromolar affinity for heme, as measured by tryptophan fluorescence quenching. The 1:1 stoichiometry of heme binding was not altered either; however, the off rate for removal of heme from the pocket increased by 2.7-fold. This may be explained by considering that loss of a heme-iron ligand weakens heme binding and may also destabilize the distal loop, allowing an increased level of solvation of the heme in the pocket. In contrast, the M362L substitution causes a decrease in the first-order rate constant for the transfer of heme to IsdA-N1 by half, from 81 to 34 s–1. The combined increase in the heme off rate by the M362L variant paired with a decreased rate of transfer to IsdA-N1 results in a ratio of catalyzed heme transfer to heme release of 6.4-fold less for the M362L variant (transfer rate ∼2600-fold greater than the off rate) than the wild-type protein (transfer rate ∼17000-fold greater than the off rate). The implication is that Met362 plays a role in the passage of heme between the IsdA and IsdB NEAT domains. The transfer of heme between the NEAT domain-containing hemophore IsdX1 and cell wall-anchored IsdC of Bacillus anthracis was observed to be biphasic and slightly slower than that seen here (fast phase of 13 s–1) but once again was at least 10000-fold faster than the observed off rate.47 The mechanism of inter-NEAT domain heme transfer is not yet known; however, in the case of IsdB-N2, Met362 may act to pull the heme-iron away from Tyr440, weakening the bond and thus facilitating transfer to IsdA. The observed bond lengthening between Tyr440 and the heme-iron in chains A and D, where Met362 mainly coordinates, supports this hypothesis.
A comparison with the streptococcal protein Shp again reveals surprising parallels. As previously mentioned, Shp uses two methionines for heme-iron coordination, namely, Met66 and Met153, located in positions structurally analogous to IsdB-N2 Met362 and Tyr440, respectively.39 Mutation of the Met66 or Met153 heme-iron ligand to Ala was found to have dramatically different effects on the heme binding and kinetic parameters of the protein.48 An M66A variant caused minimal change in the heme dissociation constant (22 μM) compared with the wild-type protein (22 μM); conversely, mutation of the other heme-iron ligand, Met153, resulted in a 3-fold increase in the heme dissociation constant (62 μM).48 An investigation of the effect of Met153 and Met66 mutation on rates of transfer to streptococcal HtsA, the cognate lipoprotein receptor for Shp-heme, also revealed striking differences between the two variants. Whereas mutation of Met66 to Ala resulted in a 7.5-fold decrease in the rate-limiting transfer step (0.4 s–1, vs 2.9 s–1 for the wild type), mutation of Met153 to Ala resulted in very little change in the rate-limiting transfer step (2.5 s–1).48 Parallels between IsdB-N2 and Shp reveal that abolishing a heme-iron ligand can have highly variable effects on the heme binding and transfer characteristics of the protein.
Nonetheless, the extent of the effects of the loss of Met362 as a heme iron ligand in IsdB is likely not yet fully characterized. As ferric iron (Fe3+) displays a preference for the phenolate of Tyr and ferrous iron (Fe2+) prefers Met or His, possessing both Met and Tyr heme-iron ligands may provide IsdB the flexibility to bind heme-iron in both oxidation states. For instance, S. aureus secretes hemolysins during infection that lyse red blood cells, releasing the stored Hb in a reduced state.49 Hb heme-iron quickly oxidizes in the bloodstream and is subsequently lost from Hb at an accelerated rate.49 If heme-iron is not completely oxidized, being able to bind ferrous heme-iron through Met362 may be advantageous for S. aureus. It is also possible that Met362 plays a role in extraction of heme from Hb; however, we have found that mixing IsdB-N2 with Hb did not result in observable heme transfer (data not shown), which is consistent with published observations that IsdB-N2 alone does not interact with Hb.22
Though IsdB-N2 and IsdH-N3 (the heme binding NEAT domain of IsdH) are 56% identical in sequence14 and both IsdB and IsdH are known to bind Hb,17,19,50 only the loss of IsdB significantly hampers the cells’ utilization of Hb as a sole iron source.19 Furthermore, a solution of IsdH-N1 (Hb binding NEAT domain) and IsdH-N3 takes up heme from Hb at a rate slightly faster than the rate of dissociation of heme from Hb, 11 h–1,50 whereas full-length IsdB takes up heme from Hb ∼100 times faster at a rate of 0.31 s–1.21 While regions outside the individual NEAT domains may be required for efficient extraction of heme by IsdH, it is also possible that the unique Met362 residue somehow participates in Hb recognition or heme extraction and expedites the process. An M362L variant of full-length IsdB is thus planned to explore this possibility.
A binding stoichiometry of 1:1 for IsdB-N2 and IsdH-N3 was estimated from reconstituted samples by using MCD.44 Similar stoichiometry would be expected on the basis of the high degree of sequence identity (56%) and similar folds (rmsd of 0.77 Å over 110 Cα atoms) of IsdB-N2 and IsdH-N3 and binding is 1:1 in both crystal structures.12 In contrast to that with IsdB-N2, a heme titration experiment with IsdH-N3 indicated a molar ratio of 4:1 leading to a model of heme stacking in the active site to provide some biological advantage,12 although it is unclear at this time what that may be, as each NEAT domain heme pocket is built to coordinate one heme at a time. An investigation of transfer kinetics between IsdH-N3 and IsdA or IsdC may help to resolve this question.
A comparison of spectroscopic and kinetic results between full-length IsdB and IsdB-N2 reveals highly similar heme binding and transfer characteristics. Zhu et al. were able to determine an off rate of 1.3 × 10–3 s–1 for recombinant full-length IsdB by transfer of heme to apoMb, close to our value of 4.8 × 10–3 s–1.21 Transfer of heme between IsdB and IsdA also appears to be closely mirrored by the NEAT domains alone. A solution of 3 μM full-length holo-IsdB transferred heme to a solution of 30 μM full-length apo-IsdA at a rate of 114 s–1,21 whereas using 2 μM holo-IsdB-N2, we observed a rate of transfer to 30 μM apo-IsdA-N1 of ∼50 s–1. Because the buffers, temperatures, and protein concentrations of the experiments differ, these rates are not directly comparable; nonetheless, they are on the same order of magnitude, supporting the hypothesis that NEAT domains alone are sufficient for heme transfer. Finally, the close similarity in the rate of transfer to IsdA or IsdA-N1 demonstrates the likelihood that the unusual Tyr-Met heme-iron coordination observed in the NEAT domain crystal structure is recapitulated in the full-length IsdB protein.
In summary, we have defined the structural basis for heme binding by IsdB. The second NEAT domain of IsdB is structurally similar to other heme binding NEAT domains, and heme-iron is coordinated by a tyrosine residue as predicted; however, we have also shown that Met362 coordinates the heme-iron on the distal side, resulting in an unprecedented mode of heme-iron coordination. We also demonstrate that Met362 plays a role in heme trafficking into and out of the heme pocket but is dispensable for stable heme binding. Lastly, we propose that the unusual nature of the heme-iron coordination suggests the possibility of a new functional role for IsdB-N2, perhaps in extraction of heme from Hb.
Acknowledgments
We thank Cherry Mao for subcloning IsdA-N1 and Dr. D. Heinrichs (University of Western Ontario, London, ON) for providing S. aureus N315 genomic DNA and critically reading the manuscript. Portions of this research were conducted at the Stanford Synchrotron Radiation Lightsource (SSRL), a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
Glossary
Abbreviations
- NEAT
near iron transporter
- Isd
iron-responsive surface determinant
- Hb
hemoglobin
- apoMb
apomyoglobin
- IsdB-N1
N-terminal NEAT domain of IsdB
- IsdB-N2
C-terminal NEAT domain of IsdB
- IsdA-N1
IsdA NEAT domain
- IsdC-N1
IsdC NEAT domain
- IsdH-N3
C-terminal NEAT domain of IsdH
- Se-Met
selenomethionine-labeled
- SE
standard error.
Supporting Information Available
Crystal packing interface between two IsdB-N2 monomers (Figure 1). This material is available free of charge via the Internet at http://pubs.acs.org.
This work is supported by Canadian Institutes of Health Research Operating Grant MOP-102596 to M.E.P.M. C.F.M.G. and J.C.G. are supported by National Sciences and Engineering Research Council Graduate Scholarships.
Supplementary Material
References
- Foster T. J. (2009) Colonization and infection of the human host by staphylococci: Adhesion, survival and immune evasion. Vet. Dermatol. 20, 456–470. [DOI] [PubMed] [Google Scholar]
- Waness A. (2010) Revisiting Methicillin-Resistant Staphylococcus aureus Infections. Journal of Global Infectious Diseases 2, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plata K.; Rosato A. E.; Wegrzyn G. (2009) Staphylococcus aureus as an infectious agent: Overview of biochemistry and molecular genetics of its pathogenicity. Acta Biochim. Pol. 56, 597–612. [PubMed] [Google Scholar]
- Wilks A.; Burkhard K. A. (2007) Heme and virulence: How bacterial pathogens regulate, transport and utilize heme. Nat. Prod. Rep. 24, 511–522. [DOI] [PubMed] [Google Scholar]
- Weinberg E. D. (2009) Iron availability and infection. Biochim. Biophys. Acta 1790, 600–605. [DOI] [PubMed] [Google Scholar]
- Skaar E. P.; Humayun M.; Bae T.; DeBord K. L.; Schneewind O. (2004) Iron-source preference of Staphylococcus aureus infections. Science 305, 1626–1628. [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K.; Skaar E. P.; Gaspar A. H.; Humayun M.; Gornicki P.; Jelenska J.; Joachmiak A.; Missiakas D. M.; Schneewind O. (2003) Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909. [DOI] [PubMed] [Google Scholar]
- Skaar E. P.; Schneewind O. (2004) Iron-regulated surface determinants (Isd) of Staphylococcus aureus: Stealing iron from heme. Microbes Infect. 6, 390–397. [DOI] [PubMed] [Google Scholar]
- Andrade M. A.; Ciccarelli F. D.; Perez-Iratxeta C.; Bork P. (2002) NEAT: A domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. GenomeBiology 3, RESEARCH0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg J. C.; Vermeiren C. L.; Heinrichs D. E.; Murphy M. E. (2007) Haem recognition by a Staphylococcus aureus NEAT domain. Mol. Microbiol. 63, 139–149. [DOI] [PubMed] [Google Scholar]
- Sharp K. H.; Schneider S.; Cockayne A.; Paoli M. (2007) Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J. Biol. Chem. 282, 10625–10631. [DOI] [PubMed] [Google Scholar]
- Watanabe M.; Tanaka Y.; Suenaga A.; Kuroda M.; Yao M.; Watanabe N.; Arisaka F.; Ohta T.; Tanaka I.; Tsumoto K. (2008) Structural basis for multimeric heme complexation through a specific protein-heme interaction: The case of the third neat domain of IsdH from Staphylococcus aureus. J. Biol. Chem. 283, 28649–28659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluym M.; Muryoi N.; Heinrichs D. E.; Stillman M. J. (2008) Heme binding in the NEAT domains of IsdA and IsdC of Staphylococcus aureus. J. Inorg. Biochem. 102, 480–488. [DOI] [PubMed] [Google Scholar]
- Pilpa R. M.; Fadeev E. A.; Villareal V. A.; Wong M. L.; Phillips M.; Clubb R. T. (2006) Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: The human hemoglobin receptor in Staphylococcus aureus. J. Mol. Biol. 360, 435–447. [DOI] [PubMed] [Google Scholar]
- Muryoi N.; Tiedemann M. T.; Pluym M.; Cheung J.; Heinrichs D. E.; Stillman M. J. (2008) Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 283, 28125–28136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann M. T.; Muryoi N.; Heinrichs D. E.; Stillman M. J. (2008) Iron acquisition by the haem-binding Isd proteins in Staphylococcus aureus: Studies of the mechanism using magnetic circular dichroism. Biochem. Soc. Trans. 36, 1138–1143. [DOI] [PubMed] [Google Scholar]
- Dryla A.; Hoffmann B.; Gelbmann D.; Giefing C.; Hanner M.; Meinke A.; Anderson A. S.; Koppensteiner W.; Konrat R.; von Gabain A.; Nagy E. (2007) High-affinity binding of the staphylococcal HarA protein to haptoglobin and hemoglobin involves a domain with an antiparallel eight-stranded β-barrel fold. J. Bacteriol. 189, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryla A.; Gelbmann D.; von Gabain A.; Nagy E. (2003) Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol. Microbiol. 49, 37–53. [DOI] [PubMed] [Google Scholar]
- Torres V. J.; Pishchany G.; Humayun M.; Schneewind O.; Skaar E. P. (2006) Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188, 8421–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harro C.; Betts R.; Orenstein W.; Kwak E. J.; Greenberg H. E.; Onorato M. T.; Hartzel J.; Lipka J.; DiNubile M. J.; Kartsonis N. (2010) Safety and immunogenicity of a novel Staphylococcus aureus vaccine: Results from the first study of the vaccine dose range in humans. Clin. Vaccine Immunol. 17, 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Xie G.; Liu M.; Olson J. S.; Fabian M.; Dooley D. M.; Lei B. (2008) Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J. Biol. Chem. 283, 18450–18460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K.; Dedent A.; Cheng A. G.; McAdow M.; Bagnoli F.; Missiakas D. M.; Schneewind O. (2010) IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 28, 6382–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson I. S.; Rosell F. I.; Scofield M.; Mauk A. G.; Murphy M. E. (2010) Directed evolution of copper nitrite reductase to a chromogenic reductant. Protein Eng., Des. Sel. 23, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.; Belin D.; Carson M.; Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne G. D.; Standaert R. F.; Karplus P. A.; Schreiber S. L.; Clardy J. (1993) Atomic Structures of the Human Immunophilin FKBP-12 Complexes with FK506 and Rapamycin. J. Mol. Biol. 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Liu M.; Lei B. (2008) The surface protein Shr of Streptococcus pyogenes binds heme and transfers it to the streptococcal heme-binding protein Shp. BMC Microbiol. 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z.; Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Vagin A.; Teplyakov A. (1997) MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- (1994) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50, 760–763. [DOI] [PubMed] [Google Scholar]
- Lamzin V. S.; Wilson K. S. (1993) Automated refinement of protein models. Acta Crystallogr. D49, 129–147. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; Macarthur M. W.; Moss D. S.; Thornton J. M. (1993) Procheck: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA. [Google Scholar]
- Eakanunkul S.; Lukat-Rodgers G. S.; Sumithran S.; Ghosh A.; Rodgers K. R.; Dawson J. H.; Wilks A. (2005) Characterization of the periplasmic heme-binding protein shut from the heme uptake system of Shigella dysenteriae. Biochemistry 44, 13179–13191. [DOI] [PubMed] [Google Scholar]
- Stein R. A.; Wilkinson J. C.; Guyer C. A.; Staros J. V. (2001) An Analytical Approach to the Measurement of Equilibrium Binding Constants: Application to EGF Binding to EGF Receptors in Intact Cells Measured by Flow Cytometry. Biochemistry 40, 6142–6154. [DOI] [PubMed] [Google Scholar]
- Hargrove M. S.; Singleton E. W.; Quillin M. L.; Ortiz L. A.; Phillips G. N. Jr.; Olson J. S.; Mathews A. J. (1994) His64(E7) → Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. J. Biol. Chem. 269, 4207–4214. [DOI] [PubMed] [Google Scholar]
- Teale F. W. (1959) Haem-globin equilibrium studies by fluorimetry. Biochim. Biophys. Acta 35, 289–291. [DOI] [PubMed] [Google Scholar]
- Aranda R.; Worley C. E.; Liu M.; Bitto E.; Cates M. S.; Olson J. S.; Lei B.; Phillips G. N. Jr. (2007) Bis-methionyl coordination in the crystal structure of the heme-binding domain of the streptococcal cell surface protein Shp. J. Mol. Biol. 374, 374–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg J. C.; Vermeiren C. L.; Heinrichs D. E.; Murphy M. E. (2007) Heme coordination by Staphylococcus aureus IsdE. J. Biol. Chem. 282, 28815–28822. [DOI] [PubMed] [Google Scholar]
- Hargrove M. S.; Barrick D.; Olson J. S. (1996) The Association Rate Constant for Heme Binding to Globin Is Independent of Protein Structure. Biochemistry 35, 11293–11299. [DOI] [PubMed] [Google Scholar]
- Liu M.; Tanaka W. N.; Zhu H.; Xie G.; Dooley D. M.; Lei B. (2008) Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem. 283, 6668–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- Tiedemann M. T.; Muryoi N.; Heinrichs D. E.; Stillman M. J. (2009) Characterization of IsdH (NEAT domain 3) and IsdB (NEAT domain 2) in Staphylococcus aureus by magnetic circular dichroism spectroscopy and electrospray ionization mass spectrometry. J. Porphyrins Phthalocyanines 13, 1006–1016. [Google Scholar]
- Beitlich T.; Kuhnel K.; Schulze-Briese C.; Shoeman R. L.; Schlichting I. (2007) Cryoradiolytic reduction of crystalline heme proteins: Analysis by UV-Vis spectroscopy and X-ray crystallography. J. Synchrotron Radiat. 14, 11–23. [DOI] [PubMed] [Google Scholar]
- Vermeiren C. L.; Pluym M.; Mack J.; Heinrichs D. E.; Stillman M. J. (2006) Characterization of the heme binding properties of Staphylococcus aureus IsdA. Biochemistry 45, 12867–12875. [DOI] [PubMed] [Google Scholar]
- Fabian M.; Solomaha E.; Olson J. S.; Maresso A. W. (2009) Heme transfer to the bacterial cell envelope occurs via a secreted hemophore in the Gram-positive pathogen Bacillus anthracis. J. Biol. Chem. 284, 32138–32146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y.; Zhu H.; Liu M.; Fabian M.; Olson J. S.; Aranda R. t.; Phillips G. N. Jr.; Dooley D. M.; Lei B. (2007) Bis-methionine ligation to heme iron in the streptococcal cell surface protein Shp facilitates rapid hemin transfer to HtsA of the HtsABC transporter. J. Biol. Chem. 282, 31380–31388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove M. S.; Whitaker T.; Olson J. S.; Vali R. J.; Mathews A. J. (1997) Quaternary structure regulates hemin dissociation from human hemoglobin. J. Biol. Chem. 272, 17385–17389. [DOI] [PubMed] [Google Scholar]
- Pilpa R. M.; Robson S. A.; Villareal V. A.; Wong M. L.; Phillips M.; Clubb R. T. (2009) Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J. Biol. Chem. 284, 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.