Abstract
Gefitinib and erlotinib, which are epidermal growth factor receptor- (EGFR-) specific tyrosine kinase inhibitors (TKIs), are widely used as molecularly targeted drugs for non-small-cell lung cancer (NSCLC). Currently, the search for EGFR gene mutations is becoming essential for the treatment of NSCLC since these have been identified as predictive factors for drug sensitivity. On the other hand, in almost all patients responsive to EGFR-TKIs, acquired resistance is a major clinical problem. Mechanisms of acquired resistance reported in the past few years include secondary mutation of the EGFR gene, amplification of the MET gene, and overexpression of HGF; novel pharmaceutical agents are currently being developed to overcome resistance. This review focuses on these mechanisms of acquired resistance to EGFR-TKIs and discusses how they can be overcome.
1. Introduction
The epidermal growth factor receptor- (EGFR-) specific tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib are molecularly targeted drugs used for the treatment of non-small-cell lung cancer (NSCLC). In clinical trials, although response rates were approximately 10%–19%, in some cases dramatic responses have been observed soon after initiation of treatment, with this trend being particularly strong in Japanese patients, women, nonsmokers, and adenocarcinoma cases [1, 2]. In 2004, three research groups reported that the existence of the activating mutations of EGFR gene was a predictive factor for sensitivity to EGFR-TKIs [3–5]. Deletion mutations, mainly occurring around codons 746–750 in exon 19, and the substitution of leucine with arginine at codon 858 in exon 21 (L858R) comprise approximately 90% of these mutations [6]. These mutations are more prevalent in Asians, women, non smokers, and patients with adenocarcinoma, groups that match the highly gefitinib-sensitive clinical subset [6]. Many investigators have reported results from retrospective analyses of associations between EGFR gene mutations and EGFR-TKI sensitivity. These analyses indicate that approximately 70%–80% of mutation-positive cases are EGFR-TKI sensitive whereas in wild-type patients the response rate is 10%–20% [6].
In recent years, three important findings have been reported regarding EGFR gene mutations and gefitinib treatment by Asian groups. First, in the IPASS trial, gefitinib treatment was compared with carboplatin and paclitaxel combination therapy in untreated East Asian patients with advanced pulmonary adenocarcinoma who were non-smokers or former light smokers [7]. The gefitinib group had a longer progression-free survival (PFS) than the carboplatin–paclitaxel group among all patient groups (hazard ratio for progression or death, 0.74). In the subgroup of patients who were positive for EGFR gene mutations, PFS was significantly longer among those who received gefitinib than among those who received carboplatin–paclitaxel therapy (9.5 months versus 6.6 months). Additionally, two Japanese groups reported the results of Phase 3 comparative clinical trials of gefitinib treatment and combined platinum-based treatment for EGFR gene mutation-positive patients. Both the WJTOG3405 [8] and NE J002 trials [9] showed better PFS for the gefitinib group (9.2 months versus 6.3 months and 10.4 months versus 5.5 months, resp.).
Although EGFR-TKI treatment shows good response rates and PFS in NSCLC patients with EGFR gene mutations as mentioned above, acquired resistance to EGFR-TKI treatment almost always develops after a median of approximately 10 months from the initiation of treatment. To date, several major mechanisms of acquired resistance, such as secondary mutation of the EGFR gene, amplification of the MET gene, and overexpression of HGF, have been reported and advances in the development of effective pharmaceutical agents against these mechanisms are being made. EGFR gene mutations such as exon 20 insertions [10, 11] and KRAS gene mutations [12] are believed to contribute to primary resistance to EGFR-TKI treatment. This review focuses on recent findings regarding the mechanisms of acquired resistance after initial response to EGFR-TKI therapy and discusses how they can be overcome.
2. Acquired Resistance
2.1. Secondary T790M Mutation of the EGFR Gene
2.1.1. About the Secondary T790M Mutation
A secondary mutation of the EGFR gene reported in 2005 was the first mechanism of acquired resistance to EGFR-TKIs to be identified [14–16]. When threonine-to-methionine mutations in codon 790 (T790M) in exon 20 of the EGFR gene occur additively as a secondary mutation, drug resistance is observed despite the occurrence of drug-sensitive activating mutations (Figure 1(c)). Crystal structure modeling has revealed that T790 is located in the ATP-binding pocket of the catalytic region and appears to be critical for the binding of erlotinib and gefitinib. T790 is often referred to as the “gatekeeper residue.” Substitution of the threonine at this codon with a bulkier residue, such as methionine, is believed to sterically hinder the binding of these drugs. This amino acid change is not expected to interfere with ATP binding and, therefore, is not expected to alter the activity of the kinase on ligand stimulation [14]. A recent analysis showed that T790M mutations do not considerably affect the binding affinity between EGFR and EGFR-TKIs but instead increase the binding affinity between EGFR and ATP, causing a relative decrease in binding with EGFR-TKIs [17]. The authors reported that increased ATP affinity is the primary mechanism by which the T790M mutation confers drug resistance. An experiment using cell lines transfected concurrently with activating mutations and a T790M mutation also proved that resistance to gefitinib and erlotinib is evident when this mutation is present [14–16].
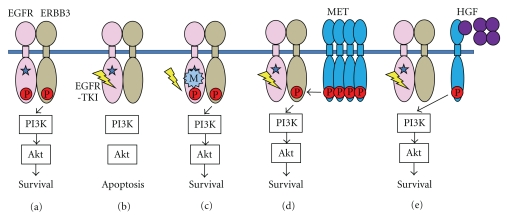
Figure 1.
Mechanism of acquired resistance to EGFR-TKI (modified from reviews of Mitsudomi and Yataba [6] and Yano [13]). (a) Survival signal through the PI3K/Akt pathway in NSCLC cells with an EGFR activating mutation (star). (b) EGFR-TKI inhibits phosphorylation of EGFR and the survival signal is shut down, leading to apoptosis of cells. (c) Secondary T790M mutation prevents binding of EGFR-TKI to EGFR, resulting in cell survival. (d) Amplified MET causes phosphorylation of ERBB3. Even when phosphorylation of EGFR is inhibited by EGFR-TKI, activation of the PI3K/Akt pathway is maintained through ERBB3. (e) HGF induces activation of the PI3K/Akt pathway through MET; this activation is independent of ERBB3 or EGFR.
Analogous secondary mutations of the BCR-ABL gene in case of chronic myelogenous leukemia (CML) [18] and the KIT gene in case of gastrointestinal stromal tumor [19] have previously been reported as mechanisms of imatinib resistance, which is also a TKI. The structural similarity between ABL and EGFR tyrosine kinases is considerably high, and T315I in ABL corresponds to T790M in EGFR [20]. Using BaF3 cells, Azam et al. revealed that a gatekeeper threonine mutation stabilizes an active conformation of these tyrosine kinases through a network of hydrophobic interactions known as the hydrophobic spine and has a transforming function [21].The authors anticipated that this regulatory mechanism would be conserved across the kinase family.
In summary, from the above three reports and subsequent reports on T790 mutations using clinical specimens from patients with acquired resistance to EGFR-TKIs [22–24], a secondary T790M mutation has been detected in approximately 50% of all patients.
2.1.2. Occurrence of T790M Mutations
Interestingly, T790M mutations exist as minor clones in almost all reported cases of acquired resistance. In every case, the results of sequence analysis or subcloning analysis indicate that the T790M mutant alleles or clones were lesser in number than wild-type alleles or clones. To address this issue, Inukai et al. proposed a clonal selection model of T790M mutant cells [25]. They used a highly sensitive method, that is, a mutant-enriched PCR assay to analyze clinical specimens from 280 sample cases who were not treated with gefitinib, and they detected 9 cases (3.6%) with a 790M mutation. Mutation was detected by direct sequencing in only one of the 9 cases; therefore the fraction of T790M mutant cells may have been considerably small. Inukai et al. hypothesized that gefitinib treatment may lead to the selection of T790M mutant cells and that even a small fraction of T790M-positive tumor cells at the beginning of treatment could lead to clinical gefitinib resistance because of selective proliferation of T790M mutant cells. Maheswaran et al. proved this hypothesis by analyzing circulating tumor cells using a highly sensitive method [26]. They analyzed pretreatment tumor samples from 26 NSCLC cases with activating EGFR gene mutations and found low levels of T790M in 10 of these cases (38%). Because detection of T790M required a relatively high number of amplification cycles, the T790M mutation has been inferred to be present in only few cells. When PFS was analyzed among these 26 cases after administering EGFR-TKI, the PFS for cases with T790M mutations during pretreatment was significantly shorter compared to that for cases without T790M mutations (median 7.7 months versus 16.5 months, P < .01). Given these results, the researchers inferred that the T790 mutant cells that existed as minor clones might be selected during treatment and become dominant after treatment.
This is analogous to the mechanism used by certain secondary mutations of the ABL genes in CML [27, 28]. Pre-existing small fractions of mutations become major clones when the tumor becomes resistant to imatinib, probably because of the selection pressure due to imatinib treatment [28].
Although rare, T790M mutations exist as major clones irrespective of gefitinib administration in certain patients [29, 30]. T790M mutations have also been reported to exist as germline mutations in families with a history of high incidence of familial lung cancer [31]. Vikis et al. reported that standalone T790M mutations exhibit increased kinase activity. Furthermore, the human bronchial epithelial cell line that transfects T790M mutations has a higher growth advantage than wild-type cells, although not as high as that of deletion mutations [32]. Mulloy et al. showed that the T790M mutant exhibits tyrosine phosphorylation levels comparable to those of wild-type EGFR, whereas the T790M/L858R double mutant exhibits a substantial increase in phosphorylation compared with the L858R mutant alone [33]. Godin-Heymann et al. also reported similar phenomena for both L858R mutations and a deletion mutation of exon 19, and discussed combination of activating mutations and T790M mutations is important to oncogenic performance [34]. Maheswaran et al. proposed that T790M may initially arise because of these oncogenic characteristics and may rapidly emerge as a dominant allele after treatment [26].
2.2. MET Gene Amplification
In 2007, a research group at the Dana-Farber Cancer Institute reported that MET gene amplification is involved in acquired resistance to gefitinib [24]. The MET gene encodes a transmembrane tyrosine kinase receptor that acts as an HGF receptor and is involved with invasion, metastasis, and angiogenesis in tumors. The authors established gefitinib-resistant clones from a sensitive cell line with EGFR exon 19 deletion mutations (HCC827) by exposing the cells to increasing concentrations of gefitinib. In these resistant cells, T790M mutations were not observed but MET gene amplification occurred. MET causes phosphorylation of ERBB3, which in turn sustains the activation of the phosphatidylinositol 3-kinase (PI3K)/Akt signal downstream. Because of these phenomena, even with gefitinib inhibiting the phosphorylation of ERBB3 by EGFR, the proliferation signal is not inhibited because of the maintenance of the phosphorylation of ERBB3 by MET, which causes the cells to become resistant to gefitinib (Figure 1(d)). Conversely, when the MET signal was suppressed in resistant cells that were dependent on MET amplification, sensitivity to gefitinib was clearly restored. In the analysis of clinical specimens from 18 cases of acquired resistance, MET gene amplification was observed in 4 cases (22%), and T790M mutations were present in 10 of 18 cases, more than half as many as were expected. The coexistence of MET gene amplification and a T790M mutation was reported in one of these cases.
Bean et al. also focused on MET gene amplification by comparing genomic profiles of EGFR mutant tumors from untreated patients with those from patients with acquired resistance using array-based comparative genomic hybridization [35]. In an analysis of 43 patients with acquired resistance, MET gene amplification was observed in 9 cases (21%). In addition, 4 of the 10 tumors with MET gene amplification had a T790M mutation. These results indicate that MET gene amplification and T790M mutation may possibly occur independently. Combining both reports, MET gene amplification is shown to be present in approximately 20% cases of acquired resistance.
MET gene amplification in cases not treated with EGFR-is said to occur with a frequency in the range of a few percent in all reports [35–38] bar one [39], where it was reported to be approximately 20%. This leads one to think of the possibility that the few cells that have undergone MET gene amplification before treatment become major clones after EGFR-TKI treatment, similar to the previously described occurrence of T790M mutations. Turke et al. performed an analysis of lung cancer patients and cell lines using the high-throughput FISH method and identified a subpopulation of cells with MET gene amplification before drug exposure [40]. Particularly in the HCC827 cell line, they discovered that the subpopulation with pre-existing MET gene amplification was rapidly selected when hepatocyte growth factor (HGF) was administered along with EGFR-TKI. In analysis of clinical specimens having acquired resistance, MET gene amplification was observed in 4 out of 27 cases (15%) (T790M mutations were observed in 15 cases, 55%). Analysis of the pretreatment specimens from those 4 cases indicated that the subpopulation with MET gene amplification was very small, less than 1% of the entire population. This proves that clonal selection occurs in the mechanism of MET gene amplification, as with the T790M mutation.
2.3. Overexpression of HGF
In 2008, a Japanese research group reported that overexpression of HGF, a specific ligand of MET, is involved in EGFR-TKI resistance [41]. They administered HGF to human adenocarcinoma cell lines that harbored EGFR exon 19 deletion mutations and showed that HGF induces resistance to gefitinib in a dose-dependent manner. Furthermore, by analyzing intracellular signal transmission pathways, HGF was shown to induce restoration of the PI3K/Akt signaling pathway through phosphorylation of MET. Interestingly, although MET gene amplification activates downstream signals by associating with ERBB3, HGF induces downstream signal activation through MET; this activation is independent of ERBB3 or EGFR (Figure 1(e)). Analysis using clinical specimens also showed cases of high HGF expression among acquired resistance cases that did not have a T790M mutation or MET amplification as well as among cases that exhibited primary resistance despite having EGFR-TKI sensitive activating EGFR gene mutations.
Turke et al. reported similar results [40]. They analyzed 27 acquired resistance cases and found that in the 16 cases for which pre- and posttreatment specimen pairs were available, HGF expression was significantly higher in the drug-resistant specimens than in the pretreatment specimens. They also found that in the 11 cases where only the specimens postresistance acquisition were available, HGF expression values were high and similar in value to those of the postresistance acquisition specimens in the paired cases. In addition to inducing EGFR-TKI standalone resistance, Yano et al. [41] found that HGF induced drug resistance by selective proliferation of clones with MET gene amplification. Although the frequency of HGF expression among cases of acquired resistance is unclear because of scarcity of reports, the frequency of coexistence of HGF expression with T790M mutations could possibly be high [42].
2.4. Other Mechanisms of Acquired Resistance
2.4.1. Other Secondary EGFR Gene Mutations
Other secondary EGFR gene mutations such as D761Y [22], L747S [43], and T854A [44] mutations have been reported. However, the frequency of all such mutations appears to be low in comparison with the T790M mutation and further analysis of such cases is required in the future.
2.4.2. IGF-1R
Guix et al. established gefitinib-resistant clones from a gefitinib-sensitive cell line of squamous cell lung carcinoma that amplified the wild-type EGFR gene (A431) by exposing the cells to increasing concentrations of gefitinib [45]. In the gefitinib-resistant cell line, hyperphosphorylation of the insulin-like growth factor-1 (IGF-1) receptor (IGF-1R) was observed instead of suppressed EGFR, and the PI3K/Akt signal was activated through IGF-1R. Gene expression analysis showed a considerably reduced expression of IGF-binding protein 3 and IGF-binding protein 4 RNA, which are known to inhibit IGF-induced activation of IGF-1R. As these analyses were performed using cell lines, results from studies conducted using clinical specimens are now required for further understanding.
3. Strategies to Overcome Acquired Resistance
3.1. Irreversible EGFR Inhibitor
Gefitinib and erlotinib are reversible EGFR-TKIs, also known as first generation EGFR-TKIs. In contrast, the drugs currently being developed to overcome resistance are second generation EGFR-TKIs, typified by irreversible EGFR-TKIs. Unlike reversible EGFR-TKIs, irreversible EGFR-TKIs covalently and irreversibly bind a cysteine residue in EGFR to the amino acid position 797 [16]. This enables them to inhibit EGFR kinase activity even in the presence of an EGFR T790M mutation. Many of these irreversible inhibitors have demonstrated activity in preclinical studies against T790M mutations. HKI-272 [46, 47] and BIBW 2992 [48, 49], which are dual inhibitors against EGFR and HER2, and PF-00299804 [50], which is a multi-inhibitor against EGFR, HER2, and HER4, are representative agents currently undergoing clinical trial.
On the other hand, these irreversible EGFR-TKIs have yet to meet expectations in terms of clinical effectiveness. All current EGFR inhibitors possess a structurally related quinazoline-based core scaffold and are identified as ATP-competitive inhibitors of wild-type EGFR. One efficacy limiting factor is believed to be the dosage limitation imposed by the toxicity because of concurrent inhibition of wild-type EGFR [46]. Zhou et al. screened an irreversible kinase inhibitor library specifically against EGFR T790M and identified a covalent pyrimidine EGFR inhibitor [51]. These agents are 30- to 100-fold more potent against EGFR T790M, and up to 100-fold less potent against wild-type EGFR, than quinazoline-based EGFR inhibitors in vitro. These mutant-selective irreversible EGFR-TKIs could be clinically more effective and better tolerated than quinazoline-based inhibitors. The results of clinical application of these agents are expected.
Cell lines resistant to irreversible EGFR-TKIs have been established using methods similar to those used for establishing reversible EGFR-TKI-resistant cell lines. Using these cell lines, the mechanisms of acquired resistance have been revealed. Ercan et al. established a resistant cell line for an irreversible EGFR-TKI by subjecting a reversible EGFR-TKI resistant cell line with a T790M mutation (established by long-term exposure of exon 19 deletion mutant PC9 cells to gefitinib) to long-term exposure to PF-00299804 [52]. In the gefitinib-resistant cell line, clones with amplified T790M mutant alleles existed in small quantities. Selective proliferation of these clones occurred on long-term exposure to PF-00299804. The researchers suggested that although irreversible EGFR-TKI may be transiently effective against cancers harboring T790M mutations, clones harboring amplified EGFR T790M will rapidly emerge in vitro and in vivo through selection of pre-existing EGFR T790M or high-expressing clones, leading to clinical drug resistance. This is similar to the mechanism of clonal selection reported for MET gene amplification [40]. In another study, Yamada et al. reported that HGF overexpression is involved not only with acquired resistance against reversible EGFR-TKIs but also with acquired resistance against irreversible EGFR-TKIs [53]. In the future, reports of further studies using clinical specimens can be expected, possibly leading to the development of pharmaceutical agents to overcome these mechanisms.
3.2. MET Inhibitors and HGF Inhibitors
The efficacy of inhibitors of MET gene amplification and HGF overexpression was revealed in a preclinical study. For MET gene amplification, the MET inhibitor PHA-665752 [24] and, for HGF overexpression, HGF-MET inhibitors, such as anti-HGF antibodies and the HGF-antagonist NK4, have been reported to be effective [54].
Establishment of resistant cell lines and the mechanism of acquired resistance against MET inhibitors have also been reported. McDermott et al. established such a cell line by continuously exposing EBC-1, an NSCLC cell line with MET amplification, to a MET inhibitor, PF2341066 [55]. In the PF2341066-resistant cell line, the EGFR signal pathways were activated. Resistance was suppressed by the combined application of PF2341066 and erlotinib.
MET gene amplification and HGF overexpression often overlap with T790M mutations. To prevent acquired resistance, combined use of an irreversible EGFR-TKI and MET inhibitors is being studied. Although tolerability issues may arise, the outlook appears promising.
4. Conclusion
Today, EGFR-TKIs are known to contribute considerably to the extension of PFS in prognosis for NSCLC. We predict that understanding and overcoming EGFR-TKI resistance mechanisms will lead to further extensions and we look forward to reviewing future analyses.
References
- 1.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. Journal of the American Medical Association. 2003;290(16):2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 2.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] Journal of Clinical Oncology. 2003;21(12):2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung, cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. New England Journal of Medicine. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Science. 2007;98(12):1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. New England Journal of Medicine. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncology. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 9.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. New England Journal of Medicine. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 10.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Medicine. 2005;2(11):p. e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clinical Cancer Research. 2008;14(15):4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Medicine. 2005;2:p. e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yano S. Studies for mechanism of drug resistance to EGFR-TKI. Gan To Kagaku Ryoho. 2010;37(8):1463–1466. [PubMed] [Google Scholar]
- 14.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. New England Journal of Medicine. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 15.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Medicine. 2005;2(3):p. e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(21):7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(6):2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105(7):2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 19.Antonescu CR, Besmer P, Guo T, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutatio. Clinical Cancer Research. 2005;11(11):4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 20.Blencke S, Ullrich A, Daub H. Mutation of threonine 766 in the epidermal growth factor receptor reveals a hotspot for resistance formation against selective tyrosine kinase inhibitors. Journal of Biological Chemistry. 2003;278(17):15435–15440. doi: 10.1074/jbc.M211158200. [DOI] [PubMed] [Google Scholar]
- 21.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nature Structural and Molecular Biology. 2008;15(10):1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clinical Cancer Research. 2006;12(21):6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 23.Kosaka T, Yatabe Y, Endoh H, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clinical Cancer Research. 2006;12(19):5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 24.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 25.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Research. 2006;66(16):7854–7858. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 26.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. New England Journal of Medicine. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100(3):1014–1018. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann WK, Komor M, Wassmann B, et al. Presence of the BCR-ABL mutation Glu255Lys prior to STI571 (imatinib) treatment in patients with Ph+ acute lymphoblastic leukemia. Blood. 2003;102(2):659–661. doi: 10.1182/blood-2002-06-1756. [DOI] [PubMed] [Google Scholar]
- 29.Toyooka S, Kiura K, Mitsudomi T, Kobayashi S, Tenen DG, Halmos B. EGFR mutation and response of lung cancer to gefitinib. New England Journal of Medicine. 2005;352(20):p. 2136. doi: 10.1056/NEJM200505193522019. [DOI] [PubMed] [Google Scholar]
- 30.Shih JY, Gow CH, Yang PC. EGFR mutation conferring primary resistance to gefitinib in non-small-cell lung cancer. New England Journal of Medicine. 2005;353(2):207–208. doi: 10.1056/NEJM200507143530217. [DOI] [PubMed] [Google Scholar]
- 31.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nature Genetics. 2005;37(12):1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 32.Vikis H, Sato M, James M, et al. EGFR-T790M is a rare lung cancer susceptibility allele with enhanced kinase activity. Cancer Research. 2007;67(10):4665–4670. doi: 10.1158/0008-5472.CAN-07-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulloy R, Ferrand A, Kim Y, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Research. 2007;67(5):2325–2330. doi: 10.1158/0008-5472.CAN-06-4293. [DOI] [PubMed] [Google Scholar]
- 34.Godin-Heymann N, Bryant I, Rivera MN, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Research. 2007;67(15):7319–7326. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okuda K, Sasaki H, Yukiue H, Yano M, Fuj Y., II Met gene copy number predicts the prognosis for completely resected non-small cell lung cancer. Cancer Science. 2008;99(11):2280–2285. doi: 10.1111/j.1349-7006.2008.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onozato R, Kosaka T, Kuwano H, Sekido Y, Yatabe Y, Mitsudomi T. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. Journal of Thoracic Oncology. 2009;4(1):5–11. doi: 10.1097/JTO.0b013e3181913e0e. [DOI] [PubMed] [Google Scholar]
- 38.Kubo T, Yamamoto H, Lockwood WW, et al. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. International Journal of Cancer. 2009;124(8):1778–1784. doi: 10.1002/ijc.24150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naïve cohort. Journal of Thoracic Oncology. 2008;3(4):331–339. doi: 10.1097/JTO.0b013e318168d9d4. [DOI] [PubMed] [Google Scholar]
- 40.Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17(1):77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yano S, Wang W, Li QI, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Research. 2008;68(22):9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 42.Onitsuka T, Uramoto H, Nose N, et al. Acquired resistance to gefitinib: the contribution of mechanisms other than the T790M, MET, and HGF status. Lung Cancer. 2010;68(2):198–203. doi: 10.1016/j.lungcan.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Medicine. 2007;4(10):1669–1680. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bean J, Riely GJ, Balak M, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clinical Cancer Research. 2008;14(22):7519–7525. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. Journal of Clinical Investigation. 2008;118(7):2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sequist LV, Besse B, Lynch TJ, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. Journal of Clinical Oncology. 2010;28(18):3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 47.Wong KK, Fracasso PM, Bukowski RM, et al. A phase I study with neratinib (HKI-272), an irreversible pan ErbB receptor tyrosine kinase inhibitor, in patients with solid tumors. Clinical Cancer Research. 2009;15(7):2552–2558. doi: 10.1158/1078-0432.CCR-08-1978. [DOI] [PubMed] [Google Scholar]
- 48.Eskens FA, Mom CH, Planting AS, et al. A phase I dose escalation study of BIBW 2992, an irreversible dual inhibitor of epidermal growth factor receptor 1 (EGFR) and 2 (HER2) tyrosine kinase in a 2-week on, 2-week off schedule in patients with advanced solid tumours. British Journal of Cancer. 2008;98(1):80–85. doi: 10.1038/sj.bjc.6604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yap TA, Vidal L, Adam J, et al. Phase I trial of the irreversible EGFR and HER2 kinase inhibitor BIBW 2992 in patients with advanced solid tumors. Journal of Clinical Oncology. 2010;28(25):3965–3972. doi: 10.1200/JCO.2009.26.7278. [DOI] [PubMed] [Google Scholar]
- 50.Engelman JA, Zejnullahu K, Gale CM, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Research. 2007;67(24):11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 51.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462(7276):1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ercan D, Zejnullahu K, Yonesaka K, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29(16):2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamada T, Matsumoto K, Wang W, et al. Hepatocyte growth factor reduces susceptibility to an irreversible epidermal growth factor receptor inhibitor in EGFR-T790M mutant lung cancer. Clinical Cancer Research. 2010;16(1):174–183. doi: 10.1158/1078-0432.CCR-09-1204. [DOI] [PubMed] [Google Scholar]
- 54.Wang W, Li Q, Yamada T, et al. Crosstalk to stromal fibroblasts induces resistance of lung cancer to epidermal growth factor receptor tyrosine kinase inhibitors. Clinical Cancer Research. 2009;15(21):6630–6638. doi: 10.1158/1078-0432.CCR-09-1001. [DOI] [PubMed] [Google Scholar]
- 55.McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Research. 2010;70(4):1625–1634. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]