Abstract
OBJECTIVE
In genome-wide association studies, performed mostly in nondiabetic individuals, genetic variability of glucokinase regulatory protein (GCKR) affects type 2 diabetes-related phenotypes, kidney function, and risk of chronic kidney disease (CKD). We tested whether GCKR variability affects type 2 diabetes or kidney-related phenotypes in newly diagnosed type 2 diabetes.
RESEARCH DESIGN AND METHODS
In 509 GAD-negative patients with newly diagnosed type 2 diabetes, we 1) genotyped six single nucleotide polymorphisms in GCKR genomic region: rs6717980, rs1049817, rs6547626, rs780094, rs2384628, and rs8731; 2) assessed clinical phenotypes, insulin sensitivity by the euglycemic insulin clamp, and β-cell function by state-of-the-art modeling of glucose/C-peptide curves during an oral glucose tolerance test; and 3) estimated glomerular filtration rate (eGFR) by the Modification of Diet in Renal Disease formula.
RESULTS
The major alleles of rs6717980 and rs2384628 were associated with reduced β-cell function (P < 0.05), with mutual additive effects of each variant (P < 0.01). The minor alleles of rs1049817 and rs6547626 and the major allele of rs780094 were associated with reduced eGFR according to a recessive model (P < 0.03), but with no mutual additive effects of the variants. Additional associations were found between rs780094 and 2-h plasma glucose (P < 0.05) and rs8731 and insulin sensitivity (P < 0.05) and triglycerides (P < 0.05).
CONCLUSIONS
Our findings are compatible with the idea that GCKR variability may play a pathogenetic role in both type 2 diabetes and CKD. Genotyping GCKR in patients with newly diagnosed type 2 diabetes might help in identifying patients at high risk for metabolic derangements or CKD.
Glucokinase regulatory protein (GCKR) is expressed in liver and, to a lesser extent, in β-cells and exerts an inhibitory effect on glucokinase (GCK) by decreasing the affinity of the latter for glucose (1). Thus, it is in a key position to regulate liver glucose balance and, perhaps, glucose-stimulated insulin secretion.
Allelic variability of two GCKR single nucleotide polymorphisms (SNPs), rs780094 and rs1260326, in strong linkage disequilibrium (LD) with each other, is associated with a number of human traits, including serum triglyceride (2–5), fasting glucose (2,5), C-reactive protein (6), coagulation protein C (7), uric acid (8), insulin levels (4), and susceptibility to type 2 diabetes (2,4,5,9,10). Moreover, the missense variant P446 L of rs1260326 is associated with reduced glomerular filtration rate (estimated glomerular filtration rate [eGFR]) and increased risk of chronic kidney disease (CKD) (11). On the whole, the major rs1260326 allele is associated with higher fasting glucose and insulin, lower eGFR, and increased CKD risk, but lower triglycerides. Thus, this single allele seems to be a multifaceted risk indicator of chronic diseases.
Most studies reporting that GCKR variation is associated with diabetes-related intermediate phenotypes were performed in nondiabetic individuals, and surrogate markers, not state-of-art methods, were used to assess β-cell function or insulin sensitivity (2,4,5,9,10). Because both traits display a significant spread in patients with type 2 diabetes, the question arises whether GCKR variants may affect β-cell function in diabetic patients, thereby potentially being a determinant of type 2 diabetes heterogeneity, a marker of the metabolic phenotype, or a tool to select optimal therapy and to better define prognosis. Moreover, because GCKR is involved in the susceptibility to CKD, it might help to select subgroups of patients with higher risk of renal complications.
We therefore undertook the present investigation to test whether the genetic variability of GCKR affects renal or β-cell function in patients with newly diagnosed type 2 diabetes. We used the database of the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS), an ongoing investigation of patients with newly diagnosed type 2 diabetes. This cohort was deemed useful for our purposes because of the absence of the potentially confounding effects of long-lasting antidiabetic treatments and of the limited impact of duration and severity of hyperglycemia per se on the metabolic phenotypes.
RESEARCH DESIGN AND METHODS
VNDS
The VNDS is an ongoing study aiming at building a biobank of patients with newly diagnosed type 2 diabetes. A detailed description of the study population and experimental design is found in the Supplementary Data. The current study reports the data collected in 509 patients whose characteristics are summarized in Table 1.
Table 1.
Clinical and metabolic features of the VNDS population
| Variable | Male | Female | All |
|---|---|---|---|
| n (M/F) | 342 | 167 | 509 |
| Age (years) | 58 [51–65] | 61 [55–66] | 59 [52–65] |
| BMI (kg/m2) | 28.7 [26.2–32.4] | 30.8 [27.4–34.5] | 29.4 [26.5–33.2] |
| Waist (cm) | 102 [94–112] | 97 [91–104] | 100 [93–108] |
| Fasting P-glucose (mmol/L) | 7 [6.2–8.0] | 7.3 [6.2–8.1] | 7.1 [6.2–8.0] |
| 2-h P-glucose (mmol/L) | 13.7 [11.0–16.4] | 13.3 [10.2–16.3] | 13.5 [10.6–16.3] |
| HbA1c (%) | 6.7 [6.1–7.6] | 6.7 [6.2–7.4] | 6.7 [6.2–7.5] |
| Triglycerides (mmol/L) | 1.4 [1.0–2.1] | 1.3 [1.0–2.0] | 1.4 [1.0–2.0] |
| HDL cholesterol (mmol/L) | 1.1 [1.0–1.3] | 1.2 [1.0–1.4] | 1.1 [1.0–1.4] |
| Cholesterol (mmol/L) | 4.9 [4.2–5.5] | 5.1 [4.5–5.8] | 5.0 [4.3–5.6] |
| Systolic blood pressure (mmHg) | 134 [120–150] | 140 [130–150] | 138 [124–150] |
| Diastolic blood pressure (mmHg) | 82 [80–90] | 85 [80–90] | 84 [80–90] |
| Insulin sensitivity (μmol/min/m2 BSA) | 565 [314–834] | 546 [394–755] | 559 [351–800] |
| Insulinogenic index (mU/mmol) | 3.5 [2.0–5.8] | 4.6 [2.3–8.1] | 3.8 [2.0–6.7] |
| CIR120’ (mU × L/mmol2) | 0.4 [0.2–1.1] | 0.6 [0.2–1.4] | 0.5 [0.2–1.2] |
| eGFR (mL/min/1.73 m2) | 84.4 [74.7–96.3] | 74.9 [62.5–86.1] | 80.0 [70.3–93.4] |
| Urinary albumin/creatinine (mg/mmol) | 0.8 [0.4–3.1] | 0.6 [0.3–1.5] | 0.8 [0.4–2.5] |
| Serum creatinine (μmol/L) | 82 [74–90] | 69 [62–80] | 79 [68–88] |
Data are presented as median [interquartile range].
Standard clinical parameters were assessed in all patients. Metabolic tests were carried out on two separate days in random order. On one day, an oral glucose tolerance test (OGTT) (75 g) was performed to assess β-cell function, as previously described (12). On a separate day, a euglycemic insulin clamp was performed to assess insulin sensitivity (13).
Analytic procedures
Plasma glucose concentration was measured in duplicate with a Beckman Glucose Analyzer II (Beckman Instruments, Fullerton, CA) at bedside. Serum C-peptide and insulin concentrations were measured by chemiluminescence as previously described. Glycated hemoglobin and serum lipids were measured by standard in-house methods. GAD antibodies were measured by immunoradiometry (CentAK, Medipan, Berlin-Dahlewitz, Germany), according to the manufacturer’s instructions. Serum creatinine was measured by Jaffe’s reaction. Urinary albumin excretion was measured in an early morning urine sample as the albumin-to-creatinine ratio by an immunonephelometric method.
Calculations
eGFR was derived from serum creatinine by the Modification of Diet in Renal Disease equation (14).
The amount of glucose metabolized during the last 60 min of the clamp (M value, reference insulin sensitivity) was computed with standard formulae (13).
The following classic indexes of β-cell function were computed (15):
Insulinogenic index: (Insulin30’ − Insulin0’)/(Glucose30’ − Glucose0’), units: mU/mmol
CIR120’: Insulin120’/[Glucose120’ * (Glucose120’ − 3.89)], units: mU * L/mmol2
in which CIR120’ is corrected insulin response at 120’. Further insights in β-cell function were sought by mathematic modeling (see below and Supplementary Data).
Modeling of β-cell function
The analysis of the glucose and C-peptide curves during the OGTTs of the VNDS follows the general strategy described in previous publications (12) and builds on previous works from other laboratories (16,17). Insulin secretion rate (ISR) is described as the sum of two components, one responding to the rate of increase of glucose and one responding to the glucose concentration itself. Modeling details are found in Supplementary Tables 1 and 2.
There are two main physiologic outputs of the model:
derivative control [pmol · m−2 BSA] · [mmol · L−1 · min−1]−1 of β-cell function, which is presented as the amount of insulin secreted in response to a rate of glucose increase of 1 mmol/L per minute that lasts for 1 min; and
proportional control of β-cell function, which is presented as the stimulus-response curve linking glucose concentration (x-axis) to ISR (pmol · min−1 · m−2 BSA; y-axis) at the preselected glucose concentrations of 5.5, 8.0, 11.0, 15.0, and 20.0 mmol/L.
Genotyping
A peripheral blood sample was collected from each patient, and the DNA was extracted by standard salting-out method.
Six tag-SNPs (rs6717980, rs1049817, rs6547626, rs780094, rs2384628, and rs8731), which represent ∼95% of the genetic variability of this genomic region (∼610 kbp) (Supplementary Fig. 1), were selected with the software GEVALT (GEnotype Visualization and ALgorithmic Tool) (18). The LD values in our population, expressed as r2, are consistent with the values reported by HapMap and are reported in Supplementary Fig. 1.
Genotypes were assessed by the high-throughput genotyping Veracode technique (Illumina Inc., San Diego, CA), applying the GoldenGate Genotyping Assay according to the manufacturer’s instructions (19).
Statistical analysis
Data are presented as medians and interquartile range, unless otherwise indicated. Hardy-Weinberg equilibrium was tested by χ2 test. Skewed variables were log-transformed to improve the approximation to the Gaussian distribution. Single variant association analyses were carried out by generalized linear models as implemented in SPSS (SPSS Inc., Chicago, IL). When statistically significant differences were found in the proportional control of β-cell function, the analyses were repeated after adjusting for a number of potential confounders, i.e., age, sex, BMI, and eGFR. Because the logarithmic transformation was insufficient to improve the distribution of the derivative control of β-cell function, the latter was first analyzed by Kruskal-Wallis test, and if statistically significant differences were found, a multivariate analysis of the log-transformed value was performed, adjusting for age, sex, BMI, and eGFR. All statistical analyses were carried out with the SPSS 12.0 software. Statistical significance was declared at P < 0.05.
RESULTS
Clinical features of the study population
Anthropometric, clinical, and metabolic features of the study subjects are shown in Table 1. Most patients were overweight or obese, but less than 50% were obese. However, abdominal obesity was present in ∼50% of male patients and in >75% of female patients. Glucose control, as measured by HbA1c, was fairly good with 25% patients having HbA1c >7.4%. High triglyceride and low HDL-cholesterol levels were common, especially the latter in the female patients. Systolic and diastolic hypertensive values were detected in 50 and 25% of the patients, respectively.
Effects of GCKR variants on β-cell and kidney functions
Allele distribution was compatible with the Hardy-Weinberg equilibrium in all six SNPs (P ≥ 0.19).
Anthropometric, clinical, and metabolic features of the study subjects, layered according to rs6717980, rs1049817, rs6547626, rs780094, rs2384628, and rs8731 alleles, are shown in Supplementary Tables 3–8.
Both rs6717980 and rs2384628 were significantly related to β-cell function. Carriers of the major (A) rs6717980 allele displayed a significant reduction in the proportional control of β-cell function, i.e., the curve relating glucose (stimulus) to ISR (response), according to a dominant model (P < 0.02, Supplementary Fig. 2). Carriers of the major (C) rs2384628 allele showed a significant impairment in both the derivative control, i.e., the response of β-cell to the rate of glucose increase (−108 ± 47.7 [pmol · m−2 BSA] · [mmol · L−1 · min−1]–1 [P < 0.03], according to an additive model [Table 2]), and the proportional control of β-cell function (P < 0.04 with a dominant model, Supplementary Fig. 2).
Table 2.
Derivative control of β-cell function in patients of the VNDS according to genotype of GCKR variants
| GCKR SNP | n | MAF | AA | AB | BB | β | SE | P value |
|---|---|---|---|---|---|---|---|---|
| rs6717980 | 487 | 0.32 | 657.6 ± 52.2 | 639.4 ± 47.0 | 643.4 ± 102.9 | 0.14 | 0.21 | 0.52 |
| rs1049817 | 486 | 0.35 | 649.6 ± 52.3 | 612 ± 48.5 | 736.5 ± 91.6 | 0.16 | 0.21 | 0.45 |
| rs6547626 | 492 | 0.34 | 645.4 ± 50.3 | 609.1 ± 49.3 | 744.2 ± 619.8 | 0.16 | 0.20 | 0.42 |
| rs780094 | 492 | 0.47 | 620.8 ± 66.3 | 634.8 ± 49.6 | 676.2 ± 57.5 | 0.19 | 0.19 | 0.33 |
| rs2384628 | 493 | 0.46 | 558.1 ± 54.3 | 651.8 ± 45.8 | 760.6 ± 88.7 | 0.43 | 0.20 | 0.03 |
| rs8731 | 483 | 0.21 | 652.9 ± 42.8 | 635.6 ± 58.8 | 716.1 ± 160.8 | −0.02 | 0.24 | 0.95 |
Data are presented as mean ± SE. Variables were log-transformed before statistical analysis to approximate a Gaussian distribution; the coefficient β and its SE refer to log-transformed values. The analysis was performed adjusting for age, sex, and BMI.
A, major allele; B, minor allele; MAF, minor allele frequency.
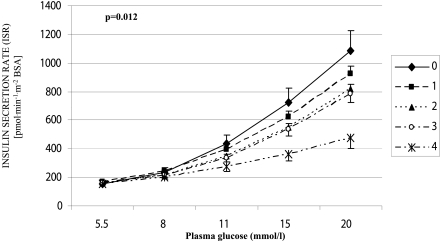
The effects of rs6717980 and rs2384628 genotypes on the proportional control of β-cell function, i.e., the curve relating glucose to ISR, were statistically independent of each other (P < 0.005 and P < 0.04, respectively, in a multivariate model including both SNPs), consistent with their low degree of LD. Therefore, we computed a GCKR score by counting one each rs6717980 A allele or each rs2384628 C allele carried by each subject. The GCKR score could range from a minimum of 0 (a genotype with neither rs6717980 A nor rs2384628 C alleles) to a maximum of 4 (a rs6717980 AA/rs2384628 CC genotype) (Supplementary Table 9). The higher the GCKR score, the lower the ISR in response to glucose (i.e., the proportional control of β-cell function) (Fig. 1, Supplementary Table 10) (unadjusted P = 0.005). The P value of the GCKR score stayed statistically significant (P = 0.006) after including in a multivariate model rs7901695 or rs7903146, the two TCF7L2 variants related to β-cell function in the VNDS (13).
Figure 1.
Effects of GCKR score on the curve relating ISR (y-axis) to glucose concentration (x-axis), i.e., the proportional control of β-cell function, in patients with newly diagnosed type 2 diabetes in the VNDS. The GCKR score was computed by counting one per each rs6717980 A allele and one per each rs2384628 C allele carried by each subject. The GCKR score could range from a minimum of 0 (a carrier of neither rs6717980 A alleles nor rs2384628 C alleles) to a maximum of 4 (a carrier of both AA in rs6717980 and CC in rs2384628). The higher the total score, the lower the β-cell insulin secretory response to glucose (unadjusted values were used for the graph; unadjusted P = 0.005; P = 0.012 after adjusting for age, sex, BMI, and eGFR). Data are presented as mean ± SEM.
Three GCKR variants were found to be related to kidney function. The minor alleles of both rs1049817 and rs6547626 (rs1049817: −3.35 ± 1.3 mL/min/1.73 m2 BSA per G allele, P = 0.01; rs6547626: −3.24 ± 1.3 mL/min/1.73 m2 BSA per A allele, P = 0.013) and the diabetes risk G allele of rs780094 (−2.56 ± 1.2 mL/min/1.73 m2 BSA per G allele, P = 0.038) were associated with reduced eGFR. Consistent with their high LD (Supplementary Fig. 1), in multivariate analysis, these three variants were not significantly associated with eGFR independently of each other (data not shown), suggesting that they may be indicators of the same genetic source of eGFR variation.
Other associations of GCKR variants to metabolic phenotypes
Carriers of the diabetes risk G allele of rs780094 had somewhat lower OGTT 2-h glucose (−0.55 ± 0.27 mmol/L, P < 0.05) (Supplementary Table 6). The major (G) allele of rs8731 was associated with increased triglycerides (+0.09 ± 0.04 log units; P < 0.04), higher systolic blood pressure (+2.66 ± 1.38 mmHg; P = 0.05), and lower insulin sensitivity (−0.1 ± 0.04 log units according to a recessive model, P < 0.04) (Supplementary Table 8). These associations stayed statistically significant after adjusting for age, sex, BMI, and eGFR.
CONCLUSIONS
The current study tested the potential role(s), if any, of six GCKR variants (Supplementary Fig. 1), purposely selected to capture ∼95% of common GCKR variability, in determining β-cell and kidney functions in patients with newly diagnosed type 2 diabetes in the VNDS. We also genotyped rs780094 because of its reported role in glucose regulation and type 2 diabetes risk (2,4,5,9,10). Novelties of our study include: 1) the use of state-of-art methods (16,17,20,21) to assess β-cell function and insulin sensitivity; and 2) the selection of the study sample, composed of patients with newly diagnosed type 2 diabetes, thereby presumably devoid of the phenotype-modifying effects of long-standing hyperglycemia or pharmacologic antidiabetic treatment.
Our data show that two (rs6717980 and rs2384628) of six GCKR SNPs exerted a strong, independent influence on β-cell function (Table 2, Supplementary Fig. 2). Indeed, homozygous carriers of “poor” β-cell function alleles at both SNPs had an ∼60% reduction in the insulin secretory response to glucose, when compared with homozygous carriers of “good” β-cell function alleles at both SNPs (Fig. 1).
This finding has three implications. First, as already shown by us for TCF7L2 (13), a role of GCKR in determining β-cell function can be detected in overt diabetes. Thus, recognizing and assessing genetic heterogeneity in patients with type 2 diabetes might turn out to be valuable in planning personalized therapies and outlining a “metabolic” prognosis. Second, despite its reported low expression in human β-cells (1), GCKR, or genomic areas closely related to it, may play a relevant role in regulating the insulin secretory response to glucose in humans (Fig. 1). Further studies will be needed to elucidate the role, if any, played by GCKR in human β-cell function at the molecular level. Third, the two SNPs associated with β-cell function in our study are in low LD with each other (Supplementary Fig. 1) and play an additive role in regulating glucose-stimulated insulin secretion (Fig. 1). Thus, multiple regions of GCKR are likely to be relevant for β-cell function in humans. Detailed genetic studies may be needed to clarify this issue.
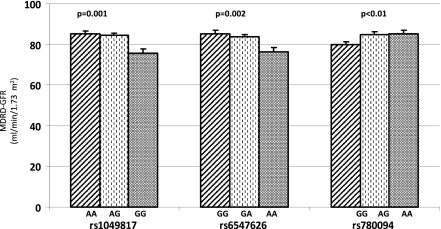
In agreement with a recently published genome-wide association study (11), three of six GCKR SNPs, in relatively high LD with each other (Supplementary Fig. 1), were significantly associated with eGFR (Fig. 2) but not to microalbuminuria (Supplementary Tables 3–8), according to an apparently recessive model. Because none of these three SNPs maintained a statistically significant association with eGFR in multivariate models with simultaneous adjustment for the other two variants, we argue that these variants may reflect the presence of one common genetic source of phenotypic variation (Fig. 2).
Figure 2.
Relationships between rs1049817, rs6547626, and rs780094 genetic variability and eGFR in patients with newly diagnosed type 2 diabetes in the VNDS. The G allele of rs1049817, the A allele of rs6547626, and the type 2 diabetes risk G allele of rs780094 were associated with reduced renal function (P ≤ 0.01), according to a recessive model. Data are presented as mean ± SEM. GFR, glomerular filtration rate; MDRD, Modification of Diet in Renal Disease.
A substantial fraction of patients with type 2 diabetes have a reduction in GFR without microalbuminuria, and, vice versa, a substantial fraction of microalbuminuric patients have normal GFR (22,23). A mild-to-moderate reduction of eGFR predicts cardiovascular disease in patients with type 2 diabetes, also independently of classic risk factors (24). Our finding that GCKR variation influences eGFR in patients with newly diagnosed type 2 diabetes supports the hypothesis of a genetic basis for nonalbuminuric CKD in this disease and, possibly, for the incremental cardiovascular risk related to this trait. Furthermore, the G allele of rs780094 is both a risk allele for type 2 diabetes (4,5,9,10) and, in this study, a marker of reduced eGFR (Fig. 2). This suggests that a genetic commonality between type 2 diabetes and nonalbuminuric CKD may exist and that the clock for CKD in these patients may start ticking long before the onset of type 2 diabetes. Although GCKR can be considered a candidate gene for glucose- and triglyceride-related (see below) phenotypes, the molecular mechanisms, if any, linking GCKR to eGFR and CKD are currently unknown.
The G allele of rs8731 was associated with increased triglycerides, increased systolic blood pressure, and reduced insulin sensitivity (Supplementary Table 8). The effect was quantitatively small and not replicated in the immediately preceding SNP; this, therefore, may be a chance finding. On the other hand, this cluster of variables belongs to the metabolic syndrome, and the quantitative magnitude (∼60 μmol/min/m2 BSA) of this putative effect of GCKR variation on whole-body insulin sensitivity is consistent with the role played by GCK, the protein regulated by GCKR, in determining insulin-mediated glucose traffic in liver. Our findings suggest a potential genetic basis for the clustering of these three traits (possibly including the cardiovascular risk they convey) in the same diabetic patient. Replication of this result in an independent cohort is needed.
Our study has a number of limitations. First, patients of the VNDS are not a population-based sample, although comparison of their features with previously published studies suggests that they are fairly representative of Italian patients with type 2 diabetes (25). However, the VNDS cohort has a somewhat higher than expected prevalence of the male sex, possibly reflecting a sex-related referral bias. Second, although we used state-of-art methods to assess β-cell function and insulin sensitivity (16,17,20,21), many relevant pathophysiologic facets were unexplored, e.g., incretin role and liver versus peripheral insulin resistance. Third, as in all association studies, our findings need to be replicated in an independent sample of patients. Fourth, our study was performed in Italian patients; extrapolations to other ethnic groups should be made with caution. Fifth, the P values we report were not adjusted for multiple comparisons, and this may result in statistical type I errors, although, in the case of GCKR, a gene well known to be implicated in the regulation of diabetes and CKD-related phenotypes, corrections for multiple comparisons might result in statistical type II errors.
In summary, we have reported that GCKR variation is significantly related in multiple ways to β-cell function and kidney function in patients with newly diagnosed type 2 diabetes. Thus, GCKR may play a polyvalent role in human disease, and assessing its variation might prove to be a useful tool to personalize the treatment and to refine the prognosis of patients with type 2 diabetes.
Supplementary Material
Acknowledgments
This study was supported in part by a European Foundation for the Study of Diabetes/Novartis grant (to R.C.B.) and research grants of the University of Verona (to R.C.B. and E.B.).
No potential conflicts of interest relevant to this article were reported.
S.B. and M.T. researched data and wrote the article. M.L.B. and F.T. researched data. G.M., E.T., P.F.P., and E.B. discussed the article. R.C.B. researched data and wrote and edited the article.
The technical help of Monica Zardini and Federica Moschetta is acknowledged (Department of Medicine, University of Verona, Verona, Italy). GCKR genotyping was performed with the XBead Reader of the Center of Functional Genomics of the University of Verona, under the supervision of Massimo Delledonne and Alberto Ferrarini, whose contribution is gratefully acknowledged.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc10-2218/-/DC1.
References
- 1.de la Iglesia N, Mukhtar M, Seoane J, Guinovart JJ, Agius L. The role of the regulatory protein of glucokinase in the glucose sensory mechanism of the hepatocyte. J Biol Chem 2000;275:10597–10603 [DOI] [PubMed] [Google Scholar]
- 2.Vaxillaire M, Cavalcanti-Proença C, Dechaume A, et al. ; DESIR Study Group The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes 2008;57:2253–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena R, Voight BF, Lyssenko V, et al. ; Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 4.Sparsø T, Andersen G, Nielsen T, et al. The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia 2008;51:70–75 [DOI] [PubMed] [Google Scholar]
- 5.Onuma H, Tabara Y, Kawamoto R, et al. The GCKR rs780094 polymorphism is associated with susceptibility of type 2 diabetes, reduced fasting plasma glucose levels, increased triglycerides levels and lower HOMA-IR in Japanese population. J Hum Genet 2010;55:600–604 [DOI] [PubMed] [Google Scholar]
- 6.Ridker PM, Pare G, Parker A, et al. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women’s Genome Health Study. Am J Hum Genet 2008;82:1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang W, Basu S, Kong X, et al. Genome-wide association study identifies novel loci for plasma levels of protein C: the ARIC study. Blood 2010;116:5032–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunnarsdóttir KA, Jensen MB, Zahrieh D, et al. CEF is superior to CMF for tumours with TOP2A aberrations: a Subpopulation Treatment Effect Pattern Plot (STEPP) analysis on Danish Breast Cancer Cooperative Group Study 89D. Breast Cancer Res Treat 2010;123:163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi Q, Wu Y, Li H, et al. Association of GCKR rs780094, alone or in combination with GCK rs1799884, with type 2 diabetes and related traits in a Han Chinese population. Diabetologia 2009;52:834–843 [DOI] [PubMed] [Google Scholar]
- 10.Bi M, Kao WH, Boerwinkle E, et al. Association of rs780094 in GCKR with metabolic traits and incident diabetes and cardiovascular disease: the ARIC Study. PLoS ONE 2010;5:e11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köttgen A, Pattaro C, Böger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet 2010;42:376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonadonna RC, Heise T, Arbet-Engels C, et al. Piragliatin (RO4389620), a novel glucokinase activator, lowers plasma glucose both in the postabsorptive state and after a glucose challenge in patients with type 2 diabetes mellitus: a mechanistic study. J Clin Endocrinol Metab 2010;95:5028–5036 [DOI] [PubMed] [Google Scholar]
- 13.Bonetti S, Trombetta M, Malerba G, et al. Variants and haplotypes of TCF7L2 are associated with {beta}-cell function in patients with newly diagnosed type 2 diabetes: the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS) 1. J Clin Endocrinol Metab 2011;96:E389–E393 [DOI] [PubMed]
- 14.Levey AS, Coresh J, Greene T, et al. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann Intern Med 2006;145:247–254 [DOI] [PubMed] [Google Scholar]
- 15.Hanson RL, Pratley RE, Bogardus C, et al. Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am J Epidemiol 2000;151:190–198 [DOI] [PubMed] [Google Scholar]
- 16.Cobelli C, Toffolo GM, Dalla Man C, et al. Assessment of beta-cell function in humans, simultaneously with insulin sensitivity and hepatic extraction, from intravenous and oral glucose tests. Am J Physiol Endocrinol Metab 2007;293:E1–E15 [DOI] [PubMed] [Google Scholar]
- 17.Mari A, Camastra S, Toschi E, et al. A model for glucose control of insulin secretion during 24 h of free living. Diabetes 2001;50(Suppl. 1):S164–S168 [DOI] [PubMed] [Google Scholar]
- 18.Davidovich O, Kimmel G, Shamir R. GEVALT: an integrated software tool for genotype analysis. BMC Bioinformatics 2007;8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin CH, Yeakley JM, McDaniel TK, Shen R. Medium- to high-throughput SNP genotyping using VeraCode microbeads. Methods Mol Biol 2009;496:129–142 [DOI] [PubMed] [Google Scholar]
- 20.Cali’ AM, Bonadonna RC, Trombetta M, Weiss R, Caprio S. Metabolic abnormalities underlying the different prediabetic phenotypes in obese adolescents. J Clin Endocrinol Metab 2008;93:1767–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss R, Caprio S, Trombetta M, Taksali SE, Tamborlane WV, Bonadonna R. Beta-cell function across the spectrum of glucose tolerance in obese youth. Diabetes 2005;54:1735–1743 [DOI] [PubMed] [Google Scholar]
- 22.Kramer HJ, Nguyen QD, Curhan G, Hsu CY. Renal insufficiency in the absence of albuminuria and retinopathy among adults with type 2 diabetes mellitus. JAMA 2003;289:3273–3277 [DOI] [PubMed] [Google Scholar]
- 23.Thomas MC, Macisaac RJ, Jerums G, et al. Nonalbuminuric renal impairment in type 2 diabetic patients and in the general population (National Evaluation of the Frequency of Renal Impairment cO-existing with NIDDM [NEFRON] 11). Diabetes Care 2009;32:1497–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ninomiya T, Perkovic V, de Galan BE, et al. ; ADVANCE Collaborative Group Albuminuria and kidney function independently predict cardiovascular and renal outcomes in diabetes. J Am Soc Nephrol 2009;20:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonora E, Targher G, Formentini G, et al. The metabolic syndrome is an independent predictor of cardiovascular disease in type 2 diabetic subjects. Prospective data from the Verona Diabetes Complications Study. Diabet Med 2004;21:52–58 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.