Abstract
Within the mammalian inner ear there are six separate sensory regions that subserve the functions of hearing and balance, although how these sensory regions become specified remains unknown. Each sensory region is populated by two cell types, the mechanosensory hair cell and the supporting cell, which are arranged in a mosaic in which each hair cell is surrounded by supporting cells. The proposed mechanism for creating the sensory mosaic is lateral inhibition mediated by the Notch signaling pathway. However, one of the Notch ligands, Jagged1 (Jag1), does not show an expression pattern wholly consistent with a role in lateral inhibition, as it marks the sensory patches from very early in their development—presumably long before cells make their final fate decisions. It has been proposed that Jag1 has a role in specifying sensory versus nonsensory epithelium within the ear [Adam, J., Myat, A., Roux, I. L., Eddison, M., Henrique, D., Ish-Horowicz, D. & Lewis, J. (1998) Development (Cambridge, U.K.) 125, 4645–4654]. Here we provide experimental evidence that Notch signaling may be involved in specifying sensory regions by showing that a dominant mouse mutant headturner (Htu) contains a missense mutation in the Jag1 gene and displays missing posterior and sometimes anterior ampullae, structures that house the sensory cristae. Htu/+ mutants also demonstrate a significant reduction in the numbers of outer hair cells in the organ of Corti. Because lateral inhibition mediated by Notch predicts that disruptions in this pathway would lead to an increase in hair cells, we believe these data indicate an earlier role for Notch within the inner ear.
The sensory patches of the inner ear offer an excellent developmental system in which to study both regional and cell fate specification (reviewed in ref. 1). Within the mammalian inner ear there are six separate sensory regions. Five of these regions perform vestibular functions, whereas just one, the organ of Corti, operates as the auditory transducer. Exactly how these sensory regions are specified remains largely unknown, although there are now several known genes that mark these areas from very early in development (2–4). The sensory regions are populated by two general cell types: the mechanosensory hair cell and the supporting cell. These cells are arranged within the epithelium in an organized and invariant fashion, such that each hair cell is surrounded by supporting cells. In the organ of Corti the pattern is even more prescribed, in that the cells are further arranged into tightly organized rows. In the mouse organ of Corti there are three rows of outer hair cells and one row of inner hair cells. The proposed mechanism for creating the sensory mosaic is lateral inhibition, in which a cell differentiating as one cell type (in this case a hair cell) prevents the surrounding cells from differentiating as the same cell type, thus creating a mosaic of cell types (5, 6). The Notch signaling pathway is known to mediate lateral inhibition in other systems (reviewed in ref. 7), and both expression analysis and perturbational experiments have supported a role for this pathway within the inner ear. For example, the Notch ligands Delta-like 1 (Dll1) and Jagged2 (Jag2) are expressed within the nascent hair cells, whereas Notch1 is initially expressed throughout the ventral wall of the cochlea before hair cell differentiation but later becomes restricted to the supporting cells (4, 8). Similarly, two members of the Hes family (Hes1 and Hes5), which are downstream effectors of Notch activation, are also expressed in the supporting cells and in the nonsensory cell types surrounding the organ of Corti (9, 10). Loss-of-function studies involving Jag2, Hes1, and Notch1 show an increased number of hair cells, the expected result of disrupting lateral inhibitory mechanisms (8, 9, 11, 12).
However, the role of one of the Notch ligands, Jagged1 (Jag1), has remained somewhat obscure, as it does not show an expression pattern wholly consistent with a role in lateral inhibition. In both chicken and mouse, Jag1 marks the sensory patches from very early in their development, perhaps indicating a role in sensory region specification (2, 4). As differentiation proceeds in the mouse, Jag1 expression is then relegated to the supporting cells, where it colocalizes with Notch1 (4). It has been proposed, based on the early expression pattern of Jag1, that it has a role in defining boundaries, such as specifying sensory versus nonsensory epithelium within the ear (2), although there has been no experimental evidence to support this hypothesis. In addition, the expression of the Notch modulator lunatic fringe (Lfng), a member of a family of molecules known to be involved in defining boundaries (reviewed in ref. 13), in domains similar to that of Jag1, lends further support to the idea that Notch signaling is involved in boundary formation in the inner ear.
Here we demonstrate that a mouse mutant, headturner (Htu), shows missing or smaller anterior and posterior ampullae, structures that house the sensory cristae. In addition there is a 33% reduction in the number of outer hair cells in the organ of Corti, although, interestingly, there is also a slight but significant increase in the number of inner hair cells and the presence of atypical hair cells. We demonstrate that Jag1 is the mutated gene responsible for these defects by identifying a missense mutation in the Jag1 gene in Htu mutants and by showing that Htu homozygotes die in utero at a similar time and because of defects similar to those of other Jag1 loss-of-function mutants.
Methods
Mice.
The Htu mutation was generated in a large-scale N-ethyl-N-nitrosourea mutagenesis program in Neuherberg, Germany (14). The Htu founder was identified because of a mild head-shaking behavior. The colony was maintained on a C3HeB/FeJ background. Embryos were taken from timed matings, and the day of the vaginal plug was considered day 0.5. Coloboma (Cm) mice were obtained from the Jackson Laboratory on a C3H/HeSnJ background. Heterozygote and wild-type Cm mice were distinguished by the presence or absence of iris colobomas at P3.
Mapping and Sequence Analysis.
Htu/+ mice on a C3HeB/FeJ genetic background were outcrossed to C57BL/6J mice, and their mutant progeny were backcrossed to C3HeB/FeJ; backcrossing to C57BL/6J led to a reduction in the penetrance of the phenotype, making classification difficult. A total of 155 offspring were obtained, of which 56 could be classified as Htu/+ by their phenotype. Only mutants that could unequivocally be phenotyped as mutant were used for mapping to avoid the false negatives' interference with the analysis. The entire genome was scanned (15), and additional chromosome 2 markers were selected for further genotyping and narrowing of the region. The ORF of Jag1 cDNA was amplified with the use of eight overlapping primer pairs (GenBank AF171092; primer sequences are available from the authors). The 5′ untranslated region was amplified with the use of the Advantage GC cDNA PCR kit (CLONTECH). Amplicons were sequenced on an ABI-377XL DNA sequencer in both the forward and reverse directions.
Genotyping.
PCR was done with the use of a forward primer at nucleotide position 1082 (5′-GTCCACGGCACCTGCAATG-3′) and a reverse primer (5′-GTGATAATGGACTGAACCTC-3′) that resides 169 bp into an intron, the preceding exon of which ends at position 1154. Because the mutation creates a FokI site, the PCR product was digested so that genotypes could be distinguished: wild types yielded a single 240-bp band; heterozygotes produced three fragments of 240 bp, 190 bp, 50 bp, and homozygotes yielded just the 190-bp and 50-bp bands.
Paintfilling.
The paintfillings of the ears were performed in a way similar to that described for embryonic chickens (16). Briefly, mice were decapitated and fixed in Bodian's fixative overnight and dehydrated in ethanol. Heads were bisected and cleared in methyl salicylate. The inner ears were injected via either the common crus and/or cochlea with the use of a pulled glass capillary pipette (20–40 μm diameter) filled with 1% gloss paint in methyl salicylate.
Scanning Electron Microscopy.
Cochleas (n = 6 Htu/+, 5 +/+, aged 20 days; n = 4 Cm/+, 2 +/+, aged 3 days) were prepared by the osmium tetroxide–thiocarbohydrazide method as described (17). For the hair cell counts, montages were made from the basal turns of the cochleas, extending between 600 and 1,200 μm for each cochlea. Counts were not performed in the apical regions because these regions were more difficult to preserve and numbers of hair cells are known to vary more. However, a qualitative examination of these areas revealed a phenotype similar to that observed in the base.
Physiology.
Mice (n = 5 Htu/+; 5 +/+; aged 60 to 83 days) were anesthetized with urethane, the middle ear was opened, and a recording electrode was placed on the round window of the cochlea. A closed sound system in the external ear canal was used to deliver calibrated tone bursts (15-ms duration, 1-ms rise/fall time, 100-ms interstimulus interval, 200 repetitions per average). Thresholds for detection of a cochlear nerve compound action potential response were obtained in 2- to 3-dB steps. The endocochlear potential was measured (n = 7 Htu/+, 5 +/+, aged 60–83 days) with a micropipette electrode inserted in the basal turn scala media through the lateral cochlear wall.
Platelet Endothelial Cell Adhesion Molecule (PECAM) Immunohistochemistry.
Embryos and yolk sacs were fixed overnight in methanol/DMSO (4:1), washed, and then incubated overnight at 4°C in anti-PECAM-1 (1:100; PharMingen). The next day embryos were washed before incubation in secondary antibody overnight at 4°C and detection the next day.
Results
Inner Ear Defects in Htu/+ Mice.
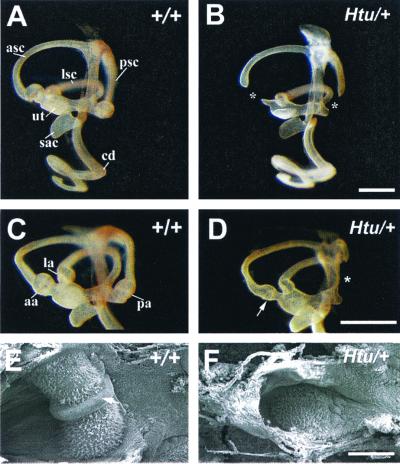
With an N-ethyl-N-nitrosourea mutagenesis approach (14) we identified a mouse mutant headturner (Htu) that showed dominant head-shaking behavior indicative of a vestibular defect. Investigation of the gross structure of the inner ear in Htu heterozygotes by paintfilling (n = 47) revealed missing posterior and sometimes anterior ampullae and a truncation of those respective semicircular canals (Fig. 1 B and D). The phenotype in which both ampullae were missing was more common (n = 29) than the phenotype where just the posterior was absent (n = 14). When present the anterior ampulla was small (Fig. 1D). In a few cases (n = 4) the posterior ampulla was present but very small. The vestibular regions (utricles and anterior and lateral cristae of six control and eight Htu/+ ears) were dissected out at and examined by scanning electron microscopy. In one mutant ear the anterior crista was present, and examination revealed that in addition to being quite small (about half the size of the control crista; Fig. 1E), the morphology of the sensory organ was also grossly abnormal. The mutant organ was much flatter than controls, and the nonsensory tissue that divides the crista (the eminentia cruciatum) was absent (Fig. 1F).
Figure 1.
Paintfilled inner ears at E16.5 showing missing or smaller ampullae in Htu/+ mutants. (A–D) Medial views of control and Htu/+ ears. (B) The phenotype where both ampullae are absent. (D) The phenotype in which just the posterior is missing. Asterisks in B and D indicate the positions of the missing ampullae. Note the smaller anterior ampulla in the Htu/+ mutant (arrow in D). (E and F) Scanning electron microscopy of the anterior crista in the control and mutant at P3. Note the small, flat appearance and the missing eminentia cruciata (arrow in E) in the mutant crista (F). aa, anterior ampulla; asc, anterior semicular canal; cd, cochlear duct; la, lateral ampulla; lsc, lateral semicircular canal; pa, posterior ampulla; psc, posterior semicircular canal; ut, utricle. (Scale bar in B = 500 μm for A and B, in D = 500 μm for C and D, and in F = 100 μm for E and F.)
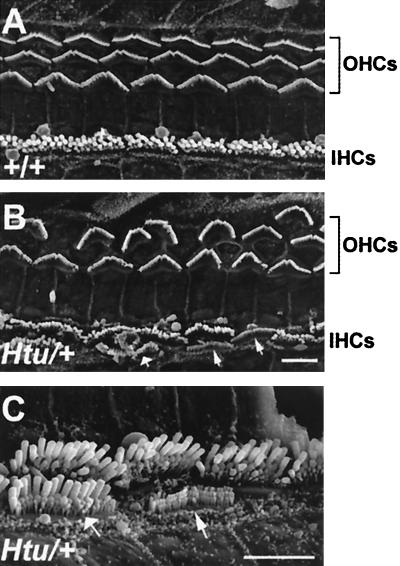
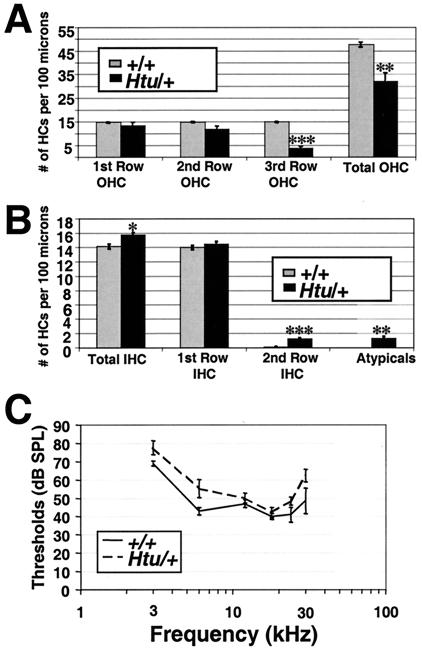
Studies of the organ of Corti in Htu heterozygotes by scanning electron microscopy at postnatal day (P) 20 revealed defects within this sensory region. In many regions of the epithelium, instead of the normal three rows of outer hair cells there were only two rows, and in some regions there was only one row (Fig. 2). Hair cell counts in the basal turn of wild-type and Htu/+ cochleas revealed a 33% reduction in the number of outer hair cells in mutants when compared with controls (Fig. 3A). Interestingly, the number of inner hair cells was increased slightly, with occasional cells appearing in a second row toward the inner sulcus. We also observed “atypical” hair cells that exhibited the bundle morphology of outer hair cells but resided in the inner hair cell row (Fig. 2C), resulting in an overall increase in the number of inner row hair cells (inners and atypicals) of 17% (Fig. 3B). Despite these patterning anomalies in the organ of Corti, Htu/+ mutants were not deaf but had slightly raised thresholds for compound action potentials, an indication of cochlear nerve responses (Fig. 3C). These differences were not significant. Endocochlear potentials were the same in the two groups (Htu/+ mean 111.0 ± 2.0 SEM; +/+ mean 112.7 ± 2.9), suggesting that the stria vascularis was functioning normally.
Figure 2.
Scanning electron micrographs of the organ of Corti in Htu/+ mice demonstrate neuroepithelial patterning defects within this organ. (A and B) Low-power views from the basal turn of the organ of Corti of a control (A) and heterozygote (B), showing the reduced number of outer hair cell rows (OHCs) in the mutant. Anomalies within the inner hair cell (IHC) row can also be observed in B and C (higher power), including extra inner hair cells (fat arrow) and atypical hair cells (thin arrows). (Scale bar in B = 5 μm for A and B. Scale bar in C = 5 μm.)
Figure 3.
Hair cell counts in the basal turn of the organ of Corti of Htu/+ mutants revealed a large decrease in the number of outer hair cells (OHC) and a smaller increase in the number of hair cells in the inner row when compared with controls. Counts varied for each cochlea but averaged approximately 1,000 μm per cochlea. (A) The number of total outer hair cells is decreased significantly, largely because of the striking loss of hair cells in the third row. (B) The total number of inner hair cells is increased slightly (but significantly) in Htu/+ mutants. This increase is due to a significant increase in the number of cells in the second row (which are seen occasionally in controls), rather than a change in the packing density, as there was no difference in the number of hair cells within the normal (first) row of inner hair cells. Atypical cells, which were never observed in control cochleas and were not included in the inner hair cell counts, occurred at about the same frequency as the extra inner hair cells (*, P < 0.05; **, P < 0.01; ***, P < 0.001; Student's two-tailed t test). (C) Compound action potential thresholds from the five littermate controls (solid line ± SEM) and the five mutants (dashed line ± SEM), aged 60–83 days, showing that thresholds were slightly higher for the mutants at all frequencies, although these differences were not significant (P > 0.05; Student's two-tailed t test).
Mapping and Sequence Analysis.
Htu was mapped to a 6.6-centimorgan region on mouse chromosome 2 between markers D2Mit399 and D2Mit280, located 60.1 to 66.7 cM, respectively, from the centromere. We considered the Jag1 gene, which also mapped to this region (18), to be a good candidate for Htu because it was known to be expressed in the sensory regions of the ear (2, 4). In addition, a study of the temporal bones of four patients with Alagille syndrome, which is caused by mutations in JAG1 (19), revealed that these patients exhibited semicircular canal defects virtually identical to those observed in the Htu/+ mice (20). As the particular combination and type of canal defects in Htu appear to be unique (with the exception of the human report), we considered this similarity to be significant.
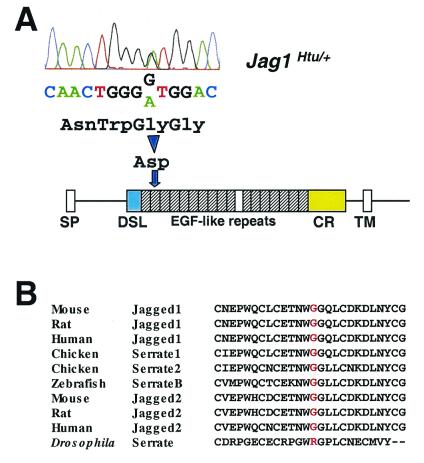
Sequence analysis of the coding region of Jag1 in Htu mutants revealed a G→A missense mutation at position 1134, resulting in a nonconservative amino acid substitution of a glycine by an aspartic acid residue (Fig. 4A). We have several reasons for proposing that this change is the causative mutation. First, on sequence comparison with other species, this glycine residue was found to be conserved in all Jagged/Serrate molecules found in vertebrates (Fig. 4B). Second, the amino acid change occurs in the extracellular domain of Jag1, in the second of the 16 epidermal growth factor (EGF)-like repeats (Fig. 4A). This second repeat, along with the first, has been shown to be important for the formation of a high-affinity complex with Notch (21). Moreover, missense mutations in the second EGF-like repeat of JAG1 have been found in patients with Alagille syndrome (22), and amino acid changes involving conserved glycine residues in the EGF-like repeats of Notch and its ligands have previously been been found to cause pathology in several different species (23–28). Finally, another mutant (slalom) derived from a separate N-ethyl-N-nitrosourea mutagenesis program (29) has been identified as having a phenotype similar to that of Htu/+ mutants and contains a mutation in the Jag1 gene (30).
Figure 4.
A missense mutation in Jag1 leads to the Htu phenotype. (A) Sequence of the Jag1 gene in a Htu/+ mouse showing the G→A mutation, the predicted protein sequence change, and its location in the schematic protein structure. The signal peptide (SP), the Delta:Serrate:LAG-2 (DSL) domain, the EGF-like repeats in the patterned boxes with the insertion sequence in a clear box, the cysteine-rich region (CR), and the transmembrane domain (TM) are shown. (B) Amino acid sequence alignment of Jagged/Serrate molecules from various species, showing the conservation of this glycine residue in vertebrates, although there is an arginine at this position in Drosophila.
Homozygote Phenotype of Htu Mutants.
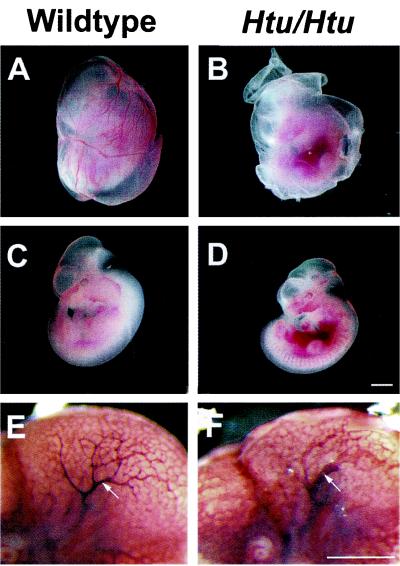
Mice that are homozygous for either an allele of the Jag1 gene in which the Delta:Serrate:LAG-2 (DSL) domain has been removed (Jag1dDSL) or homozygous coloboma (Cm) mice, which have a ≈1.1- to 2.2-cM deletion of the region encompassing the Jag1 gene (31), do not survive beyond embryonic day (E) 11.5 because of defects in vascular remodeling (18). We were therefore interested in determining whether Htu homozygotes were dying because of similar defects. Fourteen litters from Htu/+ X Htu/+ matings were examined between E10.5 and E11.5 (n = 104 embryos; 23 homozygotes). Htu/Htu embryos were identified by genotyping (see Materials and Methods) and appeared to be dying around E11.5, similar to the other Jag1 mutants. Strikingly, in all of the homozygotes examined at E11.5 there was either an absence or a great reduction in the number of large blood vessels in the yolk sacs, and the embryos themselves were hemorrhagic (Fig. 5), consistent with defects in the vascular system. Almost all of the homozygote embryos examined at E11.5 appeared to be delayed in development by a day or a half a day. In some cases an expanded pericardium or kinky neural tube was observed in the homozygote embryos (data not shown), abnormalities that were also detected in Notch mutants (32, 33). Whole-mount immunohistochemistry with an antibody recognizing a cell surface marker for endothelial cells (PECAM-1) confirmed the reduction of large blood vessels in the yolk sacs of the mutants and revealed less branching of vessels in the head region of the embryo in the mutants as compared with controls (Fig. 5 E and F).
Figure 5.
Homozygote Htu mutants die because of defects in vascular remodeling. (Left) Wild-type embryos; (Right) Htu/Htu embryos. (A and B) Yolk sacs (still containing embryos) obtained at E11.5 show the absence of large blood vessels in the homozygotes. (C and D) E11.5 embryos demonstrating the hemorrhaging observed in the mutant embryos. (E and F) E10.5 embryos processed for anti-PECAM immunohistochemistry to detect vascular defects in the embryos. Arrows indicate one of the large blood vessels in the head, which is thicker in the mutant and appears to undergo less branching in the mutant. [Scale bar = 1 mm in D (for A–D) and 500 μm in F (for E and F).]
As the homozygote phenotype was different from the heterozygote phenotype, this difference allowed a complementation test to be performed with the slalom mutant, another N-ethyl-N-nitrosourea-induced mutation that demonstrates a phenotype similar to that of Htu, and showed a different missense mutation in the second EGF-like repeat of the predicted Jag1 protein (30). Analysis of 24 embryos at E12.5 from Htu/+ × Slm/+ litters showed noncomplementation in that all five embryos that carried both mutations [confirmed by genotyping; see Materials and Methods (30)] showed an absence of blood vessels in the yolk sacs, and the embryos themselves were either necrotic (4/5) or hemorrhagic (1/5), indicating allelism.
Inner Ear Defects in Coloboma (Cm/+) Mice.
We were interested in examining known loss-of-function alleles of Jag1 to see whether they displayed inner ear defects similar to those displayed by Htu/+ mutant mice. Because Jag1 knockout mice were on a C57BL/6J background and were reported to lack behavior indicative of a vestibular defect, we considered it likely that the ear phenotype was modified in this strain because the headshaking of the Htu/+ mice was also lost with backcrossing to a C57BL/6J strain (see Materials and Methods). Therefore we examined the inner ears of Cm/+ mice, which were on a C3H background and were reported to show behavioral defects, by paintfilling and scanning electron microscopy at P3 (n = 4 Cm/+; 2 +/+). The results of this analysis revealed that all four Cm/+ ears showed anterior and posterior canal truncations near the ampullae, whereas the lateral canal was normal, and the respective ampullae were small or absent, similar to Htu/+ mutants (data not shown). A minor difference in the Cm/+ phenotype when compared with Htu/+ ears was that in two of the mutant ears the ampulla was present (although very small), but the canal was still truncated. In Htu/+ ears when the ampullae were present the canal was always complete.
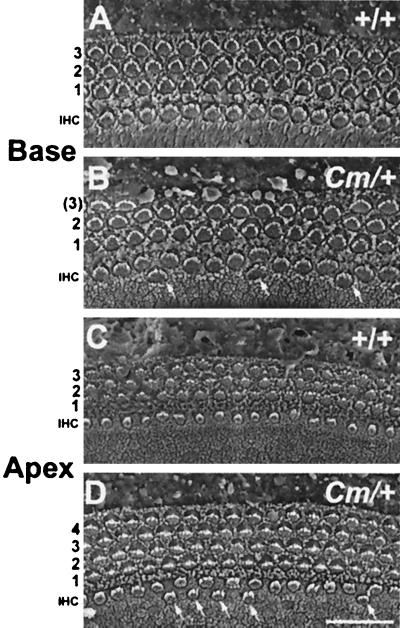
Examination of the organ of Corti of the Cm/+ mutants revealed similarities to Htu/+ mutants, but also differences. Extra hair cells in the inner hair cell row were observed in both the base and apex of Cm/+ mutants, similar to Htu/+ mutants (Fig. 6). In the basal and middle regions of the Cm/+ cochleas, regions of only two rows of OHCs were present (similar to Htu/+ mutants), but these regions occurred only occasionally and extended for short distances (5–40 μm), representing a much milder phenotype when compared with Htu/+ mutants (Fig. 6 A and B). In contrast to Htu/+ mutants, however, in the very apical 10–35% of the cochlea Cm/+ mutants showed an increase in OHC rows, demonstrating 4–5 rows of OHCs instead of the normal three (Fig. 6 C and D).
Figure 6.
Scanning electron micrographs of the organ of Corti in Cm/+ mice demonstrate some of the neuroepithelial defects observed in Htu/+ cochleas. A and B are micrographs from the basal coil of the cochlea, and C and D are taken from the apical coil of the cochlea. The position of the inner hair cells (IHC) and the number of outer hair cell rows are indicated on the left. Hair cells that are observed in a second IHC row are indicated by arrows. (Scale bar in D = 20 μm for A–D.)
Thus the similarities of the inner ear defects in Htu/+ and Cm/+ mutants provide further evidence that the missense mutation in the Jag1 gene in Htu mutants is causative. Intriguingly, the differences in the cochlear neuroepithelial defects between Htu/+ and Cm/+ indicate that the effect of the missense mutation may be more complicated than simple haploinsufficiency in some areas (see Discussion).
Discussion
Our findings indicate that Jag1 plays an important role in the specification and/or maintenance of several of the sensory organs in the inner ear, including the anterior and posterior cristae and the organ of Corti, consistent with its early expression in these sensory regions. Whether the entire ampulla is specified by Jag1 or its absence is an indirect result of loss of the crista remain to be determined. However, because Jag1 is primarily expressed in the crista and not the ampulla (4), an indirect role in ampulla development may be the more probable one. It is also unclear why the anterior and posterior crista are the most severely affected sensory organs when Jag1 is known to be expressed in all sensory regions, but it may be that these sensory organs are most sensitive to functional Jag1 levels.
Overall, the similarities of the phenotypes of Htu mutants with other presumed loss-of-function Jag1 alleles (Jag1dDSL and Cm mutants) suggest a loss-of-function effect of the missense mutation found in Htu mutants. For example, the homozygote phenotype is similar to those of both Jag1dDSL and Cm homozygote mutants, the semicircular canal phenotype is similar to that of Cm/+ mutants, and the patches of only two rows of OHCs and extra hair cells in the inner row are similar to the phenotype observed in the basal and middle turns of the Cm/+ cochlea. Indeed, because the second EGF-like repeat of Jag1 has been shown to be important for a high-affinity complex with the Notch receptor (21), a mutation in this region of the protein may destabilize ligand/receptor binding. However, the fact that the phenotype in the basal and middle turns of the organ of Corti in Cm/+ mutants is much milder in terms of OHC numbers than the phenotype in the Htu/+ organ of organ of Corti indicates that the Htu mutation may cause a dominant-negative effect in this context. In support of this possibility, another missense mutation in the second EGF-like domain of another Notch ligand, zebrafish deltaA, has also been reported to act in a dominant-negative manner (34, 35).
The presence of extra rows of OHC in the apex of Cm/+ mutant cochleas is intriguing, as it is not observed in Htu/+ mutant cochleas. Interestingly, it has been reported that using antisense oligonucleotides directed against Jag1 mRNA in cultures results in an increase in both inner and outer hair cell rows in the organ of Corti (12). One possibility is that Jag1 may have two roles: an early one, in which Jag1 specifies or maintains the sensory organs, and a later one, in which it plays a role in lateral inhibition [perhaps by increasing activation of Notch in the supporting cells (36)]. Indeed, its expression pattern in the ear suggests the possibility of two roles because it changes from being expressed throughout the entire sensory region early in development to just the supporting cells during hair cell differentiation (2, 4). In the antisense experiments Jag1 levels were only being reduced around the time of differentiation (E18-P0 rat) and thus may only be affecting the second putative role of Jag1 in lateral inhibition. Similarly, early in development the apical regions of the cochlea may be less sensitive to a change in Jag1 levels, and therefore it may be that only the second Jag1 role is being affected in the milder Cm/+ allele. A dual role for Jag1 may also explain why we observe an increase in the number of hair cells in the inner hair cell row of Htu/+ and Cm/+ mutants, as this increase may also reflect interference with a later function of Jag1. Alternatively, the organ of Corti phenotype may be explained by a failure to maintain borders on both the inner and outer sides, resulting for unknown reasons in an expansion of the hair cell region at one border (the inner side) and a decrease (or sometimes an increase) in the hair cell region at the other (outer) border. A previous study has already shown that these two boundaries may be controlled in different ways: a double knockout of Jag2/Lfng suppressed the effects of the Jag2 knockout on the expansion of the inner hair cell row but not the outer hair cell row (11). It is important to keep in mind, however, that the phenotype of the Cm/+ inner ear could be complicated by the loss of another gene(s), as at least one other gene known to be deleted in the Cm mutation (Snap25) is expressed in the ear (37). Furthermore, the genetic backgrounds of the Cm and Htu mutants are slightly different, and this difference may contribute to some of the differences observed between the two phenotypes. Therefore it will be important to compare the Htu/+ mutants to the Jag1dDSL/+ mutants on a similar genetic background.
Our results provide experimental evidence of a role for Notch signaling in the inner ear other than its known role in mediating lateral inhibition. Based on the phenotype of Htu/+ and Cm/+ mutants, the known expression pattern of Jag1 (2, 4), and the expression in the sensory patches of a molecule known to be important in boundaries (Lfng) (3), we suggest that Notch signaling may be important in the creation or maintenance of sensory/nonsensory boundaries in the inner ear.
Acknowledgments
This work was performed in partial fulfillment of the requirements for a Ph.D. degree by N.A., Sackler Faculty of Medicine, Tel-Aviv University, Israel. We thank Rick Libby and Steve Brown for helpful discussions regarding the manuscript. In addition, we thank Stavit Biton for PCR optimization with the Jag1 primers, Sarah Vreugde and Naam Kaariv for help with the mice, Orit Ben-David for help with the embryo dissections, and Charlotte Rhodes for helping with the PECAM staining. This work was supported by the Medical Research Council, Defeating Deafness, the European Commission (Contract BMH4-CT97-2715), and the German Human Genome Project.
Abbreviations
- PECAM-1
platelet endothelial cell adhesion molecule-1
- EGF
epidermal growth factor
- DSL
Delta:Serrate:LAG-2
- E
embryonic day
- OHC
outer hair cell
- Cm
coloboma
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fekete D M. Curr Opin Neurobiol. 1996;6:533–541. doi: 10.1016/s0959-4388(96)80061-4. [DOI] [PubMed] [Google Scholar]
- 2.Adam J, Myat A, Roux I L, Eddison M, Henrique D, Ish-Horowicz D, Lewis J. Development (Cambridge, UK) 1998;125:4645–4654. doi: 10.1242/dev.125.23.4645. [DOI] [PubMed] [Google Scholar]
- 3.Morsli H, Choo D, Ryan A, Johnson R, Wu D K. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison A, Hodgetts C, Gossler A, Hrabé de Angelis M, Lewis J. Mech Dev. 1999;84:169–172. doi: 10.1016/s0925-4773(99)00066-0. [DOI] [PubMed] [Google Scholar]
- 5.Lewis J. In: Regeneration of Vertebrate Sensory Cells. Bock G R, Whelan J, editors. Chichester: Wiley; 1991. pp. 25–53. [Google Scholar]
- 6.Corwin J T, Jones J E, Katayama A, Kelley M W, Warchol M E. In: Regeneration of Vertebrate Sensory Cells. Bock G R, Whelan J, editors. Chichester: Wiley; 1991. pp. 103–130. [Google Scholar]
- 7.Artavanis-Tsakonas S, Rand M D, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 8.Lanford P J, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley M W. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- 9.Zheng J L, Shou J, Guillemot F, Kageyama R, Gao W. Development (Cambridge, UK) 2000;127:4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- 10.Lanford P J, Shailam R, Norton C, Gridley T, Kelley M W. J Assoc Res Otolaryngol. 2000;1:161–171. doi: 10.1007/s101620010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang N, Martin G V, Kelley M W, Gridley T. Curr Biol. 2000;10:659–662. doi: 10.1016/s0960-9822(00)00522-4. [DOI] [PubMed] [Google Scholar]
- 12.Zine A, Van De Water T R, de Ribaupierre F. Development (Cambridge, UK) 2000;127:3373–3383. doi: 10.1242/dev.127.15.3373. [DOI] [PubMed] [Google Scholar]
- 13.Irvine K D. Curr Opin Genet Dev. 1999;9:434–441. doi: 10.1016/S0959-437X(99)80066-5. [DOI] [PubMed] [Google Scholar]
- 14.Hrabé de Angelis M, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, et al. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 15.Kiernan A E, Zalzman M, Fuchs H, Hrabe de Angelis M, Balling R, Steel K P, Avraham K B. J Neurocytol. 1999;28:969–985. doi: 10.1023/a:1007090626294. [DOI] [PubMed] [Google Scholar]
- 16.Bissonnette J P, Fekete D M. J Comp Neurol. 1996;368:620–630. doi: 10.1002/(SICI)1096-9861(19960513)368:4<620::AID-CNE12>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Self T, Mahony M, Fleming J, Walsh J, Brown S D, Steel K P. Development (Cambridge, UK) 1998;125:557–566. doi: 10.1242/dev.125.4.557. [DOI] [PubMed] [Google Scholar]
- 18.Xue Y, Gao X, Lindsell C E, Norton C R, Chang B, Hicks C, Gendron-Maguire M, Rand E B, Weinmaster G, Gridley T. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- 19.Oda T, Elkahloun A G, Pike B L, Okajima K, Krantz I D, Genin A, Piccoli D A, Meltzer P S, Spinner N B, Collins F S, et al. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 20.Okuno T, Takahashi H, Shibahara Y, Hashida Y, Sando I. Arch Otolaryngol Head Neck Surg. 1990;116:217–220. doi: 10.1001/archotol.1990.01870020093025. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, Hamada Y, Yazaki Y, Hirai H. J Biol Chem. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- 22.Crosnier C, Driancourt C, Raynaud N, Dhorne-Pollet S, Pollet N, Bernard O, Hadchouel M, Meunier-Rotival M. Gastroenterology. 1999;116:1141–1148. doi: 10.1016/s0016-5085(99)70017-x. [DOI] [PubMed] [Google Scholar]
- 23.Eldadah Z A, Hamosh A, Biery N J, Montgomery R A, Duke M, Elkins R, Dietz H C. Hum Mol Genet. 2001;10:163–169. doi: 10.1093/hmg/10.2.163. [DOI] [PubMed] [Google Scholar]
- 24.Sidow A, Bulotsky M S, Kerrebrock A W, Bronson R T, Daly M J, Reeve M P, Hawkins T L, Birren B W, Jaenisch R, Lander E S. Nature (London) 1997;389:722–725. doi: 10.1038/39587. [DOI] [PubMed] [Google Scholar]
- 25.Tax F E, Yeargers J J, Thomas J H. Nature (London) 1994;368:150–154. doi: 10.1038/368150a0. [DOI] [PubMed] [Google Scholar]
- 26.Kelley M R, Kidd S, Deutsch W A, Young M W. Cell. 1987;51:539–548. doi: 10.1016/0092-8674(87)90123-1. [DOI] [PubMed] [Google Scholar]
- 27.Lieber T, Wesley C S, Alcamo E, Hassel B, Krane J F, Campos-Ortega J A, Young M W. Neuron. 1992;9:847–859. doi: 10.1016/0896-6273(92)90238-9. [DOI] [PubMed] [Google Scholar]
- 28.Xu T, Caron L A, Fehon R G, Artavanis-Tsakonas S. Development (Cambridge, UK) 1992;115:913–922. doi: 10.1242/dev.115.4.913. [DOI] [PubMed] [Google Scholar]
- 29.Nolan P M, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray I C, Vizor L, Brooker D, Whitehill E, et al. Nat Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 30.Tsai H, Hardisty R, Rhodes C, Kiernan A E, Roby P, Tymowska-Lalanne Z, Mburu P, Rastan S, Hunter A J, Brown S D M, et al. Hum Mol Genet. 2000;10:507–512. doi: 10.1093/hmg/10.5.507. [DOI] [PubMed] [Google Scholar]
- 31.Hess E J, Collins K A, Copeland N G, Jenkins N A, Wilson M C. Genomics. 1994;21:257–261. doi: 10.1006/geno.1994.1254. [DOI] [PubMed] [Google Scholar]
- 32.Huppert S S, Le A, Schroeter E H, Mumm J S, Saxena M T, Milner L A, Kopan R. Nature (London) 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 33.Krebs L T, Xue Y, Norton C R, Shutter J R, Maguire M, Sundberg J P, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 34.Appel B, Fritz A, Westerfield M, Grunwald D J, Eisen J S, Riley B B. Curr Biol. 1999;9:247–256. doi: 10.1016/s0960-9822(99)80113-4. [DOI] [PubMed] [Google Scholar]
- 35.Riley B B, Chiang M, Farmer L, Heck R. Development (Cambridge, UK) 1999;126:5669–5678. doi: 10.1242/dev.126.24.5669. [DOI] [PubMed] [Google Scholar]
- 36.Eddison M, Le Roux I, Lewis J. Proc Natl Acad Sci USA. 2000;97:11692–11699. doi: 10.1073/pnas.97.22.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safieddine S, Wenthold R J. Eur J Neurosci. 1999;11:803–812. doi: 10.1046/j.1460-9568.1999.00487.x. [DOI] [PubMed] [Google Scholar]