Abstract
Colonization of the gastrointestinal tract and composition of the microbiota may be influenced by components of the diet, including trace elements. To understand how selenium regulates the intestinal microflora, we used high-throughput sequencing to examine the composition of gut microbiota of mice maintained on selenium-deficient, selenium-sufficient, and selenium-enriched diets. The microbiota diversity increased as a result of selenium in the diet. Specific phylotypes showed differential effects of selenium, even within a genus, implying that selenium had unique effects across microbial taxa. Conventionalized germ-free mice subjected to selenium diets gave similar results and showed an increased diversity of the bacterial population in animals fed with higher levels of selenium. Germ-free mice fed selenium diets modified their selenoproteome expression similar to control mice but showed higher levels and activity of glutathione peroxidase 1 and methionine-R-sulfoxide reductase 1 in the liver, suggesting partial sequestration of selenium by the gut microorganisms, limiting its availability for the host. These changes in the selenium status were independent of the levels of other trace elements. The data show that dietary selenium affects both composition of the intestinal microflora and colonization of the gastrointestinal tract, which, in turn, influence the host selenium status and selenoproteome expression.—Kasaikina, M. V., Kravtsova, M. A., Lee, B. C., Seravalli, J., Peterson, D. A., Walter, J., Legge, R., Benson, A. K., Hatfield, D. L., Gladyshev, V. N. Dietary selenium affects host selenoproteome expression by influencing the gut microbiota.
Keywords: selenoproteins, germ-free mice, high-throughput sequencing
Intestinal microorganisms play an important role in human physiology by regulating such processes as maturation and proliferation of the intestinal cells, food digestion, protection from pathogenic bacteria, and modulation of the mucosal immune response (1, 2). Growing evidence also links imbalances in the composition of the gut microbiota to complex diseases, including colon cancer, obesity, inflammatory bowel disease, and Crohn's disease (3–8), suggesting that dietary interventions affecting the gut microbiota hold substantial promise as alternative approaches to influence human health (9). One of the main strategies to regulate the gut microbiota through diet is the use of prebiotics, which are defined as nondigestible food ingredients that beneficially affect the host by selectively stimulating the growth and/or activity of one or a limited number of bacterial species in the colon (10). Because prebiotics promote growth of a select group of microorganisms, other strategies must also be examined, including those that can selectively inhibit or promote growth of species associated with complex diseases.
One such strategy may involve modulation of dietary trace elements. Gut microorganisms, like their hosts, are expected to be sensitive to trace elements. Some species require trace elements such as selenium (Se) for normal metabolic functions, whereas these same elements can be toxic to other microorganisms, even at very low concentrations (11, 12). Therefore, changes in dietary trace elements may modulate the composition of intestinal microbiota. For example, reducing dietary iron (Fe) caused an increase in the growth of certain organisms in the small intestine, including anaerobes, microaerophiles, lactobacilli, and enterococci. On the other hand, an increase in dietary Fe suppressed anaerobes, presumably due to elevated oxidative stress (13). However, the effect of other trace elements on intestinal microbiome is largely unknown.
Se is an essential trace element that plays an important role in human health. In particular, it is required for biosynthesis of selenoproteins, which participate in the regulation of cellular redox homeostasis, protection from oxidative stress, immune response, cancer chemoprevention, and other processes (14). Supplemental Se has been shown to be effective in decreasing incidence and mortality from several forms of cancer, including colon cancer, in both mouse models and humans (15). On the other hand, some selenoproteins may promote carcinogenesis, so the role of Se in cancer development is complex.
We hypothesized that Se supplementation may modify composition of mouse microbiota. At the same time, microflora may sequester Se, thus limiting its availability to the host (16). A better understanding of these interactions might reveal novel aspects of the influence of the microbiota on metabolism of the host. The purpose of the current study was to characterize the impact of Se diets on microbiota and the role of microbiota in influencing Se status of the host.
MATERIALS AND METHODS
Animals and feeding protocols
The diets used in the study, including 0-, 0.1-, 0.4-, and 2.25-μg/g (ppm) Se diets, were purchased from Harland TekLad (Madison, WI, USA). As described previously (17), these diets are based on the Torula yeast Se-deficient diet. Se was provided in the form of sodium selenite; 0, 0.1, 0.4, and 2.25 ppm Se indicate the added amounts of Se. In the first experiment, 4-wk-old C57BL/6J male mice (purchased from Jackson Laboratory, Bar Harbor, ME, USA) were subjected to these 4 Se diets (8 mice/group). Ten weeks later, feces were collected and frozen until further analysis.
In the second experiment, 10-wk-old germ-free Swiss Webster male mice were subjected to 3 diets: 0, 0.1, and 0.4 ppm Se. One cohort of these animals (5 mice/group) was maintained germ free, and these mice are designated germ-free (GF) mice. Another group (5 mice/group) was placed in a similar germ-free environment, but it was conventionalized with the microflora prepared from the intestine of C57BL/6J mice used in the previous experiment. These mice are designated as conventionalized (CV) mice. Both groups of animals were maintained on the same Se diets, in the same facility and at the same time, under specific pathogen-free conditions with 12-h light-dark cycle. All animal experiments were approved by the University of Nebraska–Lincoln (UNL) Institutional Animal Care and Use Committee.
Sample preparation
After 8 wk on the diets, feces from CV mice were collected and frozen until further analysis. Then, GF and CV mice were sacrificed; their tissues were collected and immediately frozen in liquid nitrogen. For Western blot analysis and activity measurements, tissues were homogenized in PBS supplemented with protease inhibitor cocktail (Sigma, St. Louis, MO, USA). Cellular debris was removed by centrifugation at 13,000 rpm for 15 min at 4°C, and protein concentration was determined with the Bradford assay.
DNA isolation and pyrosequencing
Total DNA was extracted from the fecal pellets of 8 mice/feeding group, as described previously (18). The 16S rRNA gene was amplified using modified F8 and R357 universal primers. These 16S rRNA PCR products were then subjected to pyrosequencing on a Roche/454 GS-FLX instrument (Roche, Basel, Switzerland) in the UNL Core for Applied Genomics and Ecology. The PCR products from each animal were bar coded and then mixed into a single sequencing run on the machine, so as to generate ∼2000 reads/animal. Taxonomy-dependent analysis was first performed using the naive bayesian Classifier algorithm of the Ribosomal Database Project (RDP; Michigan State University, East Lansing, MI, USA; ref. 19). Phylotypes were determined at 97% cutoffs using CD-Hit, and representative sequences were searched against the RDP database using SeqMatch. Statistical analyses were performed on taxonomic groups from Classifier and CD-Hit phylotypes by normalizing the number of reads per taxon or phylotype by the total number of reads for a given animal. These normalized proportions were then tested for statistical significance by ANOVA. For rarefaction analysis, phylotypes were assigned using the RDP pipeline (Aligner+ complete linkage clustering) and rarified using the Web-based rarefaction tools.
Western blot analyses and activity assays
Expression of selenoproteins glutathione peroxidase 1 (GPx1), selenoprotein P (SelP), and methionine-R-sulfoxide reductase 1 (MsrB1) was analyzed by Western blots with polyclonal antibodies specific for these proteins. To assay for GPx1 activity, a glutathione peroxidase kit was used (Sigma). MsrB activity was measured in an HPLC assay. Briefly, 200 μg of total protein was added to a reaction mixture (100 μl), the mixture was kept at 37°C for 30 min in the presence of 20 mM DTT, and 200 μM dabsyl-methionine-R-sulfoxide was added. After stopping the reaction by adding 200 μl acetonitrile, it was centrifuged at 4°C for 15 min at 13,000 rpm, and the supernatant (50 μl) was injected onto a C18 column (Zorbax Eclipse XDB-C18; Agilent Technologies, Santa Clara, CA, USA) to quantify the resulting dabsylated methionine.
RNA isolation and quantitative PCR
To analyze for SelP mRNA expression in the liver, total RNA was isolated by TriZOL extraction (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. Genomic DNA was removed using a DNA removal kit (Ambion, Austin, TX, USA). RNA concentration was measured spectrophotometrically, and cDNA was obtained with SuperScript III Reverse Transcriptase (Invitrogen). Real time PCR was performed using a Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). Primer sequences for the SelP expression analysis were as described previously (20): 5′-CCTTGGTTTGCCTTACTCCTTCC-3′ and 5′-TTGTTGTGGTGTTTGTGGTGG-3′. SelP mRNA expression level was normalized to that of actin mRNA.
Analyses of trace elements
To measure Se content in biological materials, we applied quantitative inductively coupled plasma mass spectroscopy (ICP-MS). Tissue extracts were digested with the mixture of 15% nitric acid and 15% hydrogen peroxide for 2 h at 70°C. After digestion, samples were diluted 10× with PBS, and the internal standard (gallium) was added to a final concentration of 50 ppb. Each sample was analyzed 3 times and in triplicate. The ICP-MS analysis was performed at the UNL Redox Biology Center spectroscopy core facility, using 7500 Agilent Technologies and Elemental Scientific (Omaha, NE, USA) SC4 autosampler instruments operating with a collision chamber with 3.5 ml H2/min and 1.5 ml He/min to eliminate polyatomic interferences on the 78Se counts. Data were normalized to protein concentration in each sample.
RESULTS
Dietary Se regulates composition of the intestinal microbiota
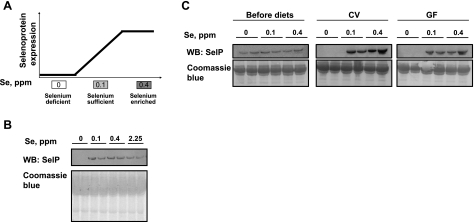
To assess the effect of dietary Se on the composition of mouse microbiota, 2-mo-old male C57BL/6J mice were subjected to a Se-deficient diet and diets supplemented with 0.1, 0.4, or 2.25 ppm of Se in the form of sodium selenite. Previously, these diets were shown to modulate selenoprotein expression (17). The expected (i.e., based on what is known about regulation of selenoprotein expression) changes in selenoprotein levels are shown schematically in Fig. 1A. After 8 wk on the diets, the Se status of mice was examined by analyzing the levels of SelP, the main selenoprotein in plasma of mammals. SelP decreased dramatically in mice fed the Se-deficient diet, whereas, as expected, the differences in SelP levels among mice fed other diets were minimal (Fig. 1B).
Figure 1.
Mouse models for examining the interrelationships between dietary Se, gut microbiota, and host Se status. A) Schematic illustration of selenoprotein expression in mice subjected to Se-deficient (0 ppm Se), Se-sufficient (0.1 ppm Se), and Se-enriched (0.4 ppm Se) diets. B, C) SelP expression was analyzed by Western blotting (WB) in plasma of conventional (B) and GF and CV (C) mice fed the indicated Se diets for 8 wk (B) or 6 wk (C; top panels); gels stained with Coomassie blue served as loading controls (bottom panels); 2 mice/group were analyzed.
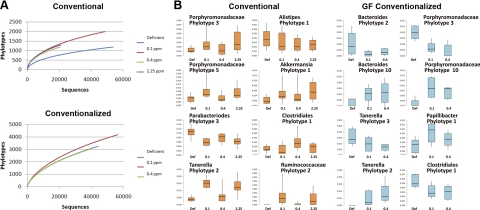
Feces from animals in the Se diet groups were collected, and the gut microbiota were examined by pyrosequencing of 16S ribosomal RNA tags at a sequencing depth of ∼2000 reads/animal. Using a threshold of 97% for phylotype assignment, rarefaction analysis of the data pooled by Se diet group showed a significant effect of Se deficiency on the numbers of phylotypes that were detected. Overall, Se in the diet increased the diversity of the microbiota (Fig. 2A). This Se-induced increase was similar at all supplementation levels. To examine the effects of Se on major taxonomic groups of the microbiota, the proportions of taxa at the phylum, class, and order level from Classifier data were analyzed by ANOVA. No significant differences were observed on the relative proportions of major taxonomic groups (data not shown). The effects on specific phylotypes were then examined using 97% CD-Hit clusters. ANOVA identified several phylotypes showing significant effects of diet (Fig. 2B). Remarkably, there were differential effects on related taxa. For example, some phylotypes belonging to the class Bacteriodales showed increases in response to Se (e.g., Porphyromonadaceae phylotypes 1 and 3, Tanerella phylotype 2), whereas others, such as Alistipes phylotype 1 and Parabacteroides phylotype 3 declined. Relative to absolute abundance, the decline in Parabacteroides in response to Se in the diet was by far the most significant effect on the microbiota. As with many phylotypes in the Bacteriodetes, several of the Firmicutes also showed modest effects in response to Se, including phylotypes within the Clostridia and Erysipelotrichi.
Figure 2.
Effect of dietary Se on gut microbiota. A) Rarefaction analysis was performed on pyrosequencing data from conventional and GF conventionalized animals. Sequences were pooled from animals of a similar feeding level and aligned using the complete linkage clustering at 97% cutoff available on the RDP pipeline, and clusters were subjected to rarefaction analysis. B) Box and whisker plots depict relative abundance of selected phylotypes showing statistically significant effects of dietary selenium (P<0.05). The taxonomic relationship of each phylotype is indicated above the relevant graph. Left panels: phylotypes from conventional animals (orange boxes). Right panels: phylotypes from GF conventionalized animals (light blue boxes). Vertical bars indicate range of proportions for each treatment group; boxes indicate upper and lower bounds of the 95% confidence intervals.
Se supplementation influences bacterial colonization of the gastrointestinal tract
To determine whether the microbiota have a significant effect on the physiological distribution of dietary Se, GF Swiss Webster mice were treated with the same 3 Se diets (0, 0.1, and 0.4 ppm Se), except that these diets were sterilized. One cohort of these animals was maintained GF, while the gut microbiota was reestablished in the other cohort using fecal materials from the CV C57BL/6J animals used in the previous experiment. Before and after 8 wk of diet treatment and conventionalization, SelP levels were examined in plasma, and fecal samples were taken at the end of the experiment and subjected to the analysis of microbiota. Western blots revealed undetectable SelP in all mice fed the Se-deficient diet, whereas little difference was observed between the 0.1- and 0.4-ppm Se groups (Fig. 1C). Thus, dietary Se affects selenoprotein expression independent of GF, CV, or colonization conditions in mice.
Similar to the conventional animals, pyrosequencing of the microbiota from conventionalized GF animals showed effects on the microbiota. Most notable was an increase in the overall diversity of the microbiota (Fig. 2A). While the 0.4-ppm diet in the conventionalized GF animals showed significantly increased diversity, the 0.1-ppm diet did not. Statistical analysis of the phylotype abundances in the conventionalized GF animals showed that several phylotypes were affected by dietary Se (Fig. 2B). In general, these phyla belonged to the same taxonomic groups as those affected in the conventional animals, including phylotypes related to Tanerella and Porphyromonadaceae. As in the conventional animals, these phylotypes showed differential effects, with some being stimulated by Se and others significantly decreasing. Likewise, one of the phylotypes belonging to the Clostridiales showed significant increases in response to Se in the conventional animals, but another phylotype was significantly reduced in response to dietary Se in the conventionalized GF animals. Collectively, our results show that dietary Se affects the overall diversity of the microbiota and has differential effects on specific taxonomic groups, even in related taxa. These effects were observed whether the microbiota was obtained naturally or by conventionalizing GF animals.
Microbial colonization affects the levels and activities of selenoproteins of the host
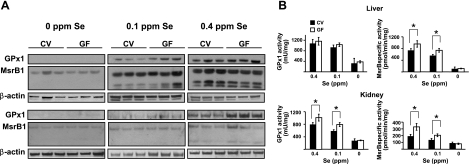
Gastrointestinal microbiota was recently shown to affect Se status and selenoprotein expression in mice (16). In that study, conventional FVN/NHanTMHsd maintained for 5 wk on Se-deficient and Se-adequate diets showed reduced GPx and/or thioredoxin reductase 1 (TR1) activities in plasma, liver, and/or intestine under Se-limiting conditions, when compared to the corresponding GF group. In our study, we analyzed the status of stress-related selenoproteins, GPx1 and MsrB1, in tissues of GF mice in comparison with those in CV mice, which served as controls, thus allowing us to directly examine the effect of gut colonization on the selenoprotein status of animals. In this experiment, GF and CV mice were subjected, in parallel, to 0-, 0.1-, and 0.4-ppm Se diets, and expression levels and activities of GPx1 and MsrB1 were analyzed in liver and kidney. Both GF and CV mice showed similar patterns of selenoprotein regulation. Their expression in mice on the 0-ppm Se diet was dramatically lower, and we also observed differences in selenoprotein expression between 0.1- and 0.4-ppm Se diets. Selenoenzyme activities paralleled the expression patterns. Total GPx activity (GPx1 is the main GPx in liver and kidney of mice; thus, this activity largely corresponds to the activity of GPx1), and the total MsrB activity (MsrB1 is the main MsrB in liver and kidney) was decreased severalfold in the 0-ppm Se group compared to the 0.1-ppm Se group, whereas the 0.4-ppm Se group showed a statistically significant increase over the 0.1-ppm Se group. This is consistent with the previous findings that expression of GPx1 and MsrB1 is maximized at ∼0.15 ppm Se in the diet (21).
However, we were particularly interested in the effect of gut colonization on the regulation of selenoprotein expression (Fig. 3). Indeed, within the 0.1-ppm Se groups, we observed higher GPx1 expression in liver and kidney, and higher GPx activity in kidneys of GF mice, compared to CV mice. In addition, within the 0.4-ppm Se groups, higher GPx1 expression and activity were observed in kidneys, and we also found higher MsrB activity in liver and kidneys of GF mice. In the 0-ppm Se group, we did not observe differences between GF and conventionalized mice, probably because of low expression and activity of selenoproteins, which approached background levels. Overall, there was a clear pattern wherein conventionalization of mice decreased expression and activity of stress-related selenoproteins in liver and kidney.
Figure 3.
Selenoprotein expression in GF and CV mice. Expression (A) and specific GPx1 and MsrB1 activities (B) in livers and kidneys are shown; 3 mice/group were analyzed. A) Expression of GPx1 and MsrB1 was analyzed by Western blots in livers (top panels) and kidneys (bottom panels) of CV and GF mice fed 0-, 0.1-, and 0.4-ppm Se diets. B) GPx and MsrB activities in liver and kidney lysates of GF and CV mice fed 0-, 0.1-, and 0.4-ppm Se diets. Solid bars, CV mice; open bars, GF mice. *P < 0.05; Student's t test.
Gut colonization and SelP levels in plasma
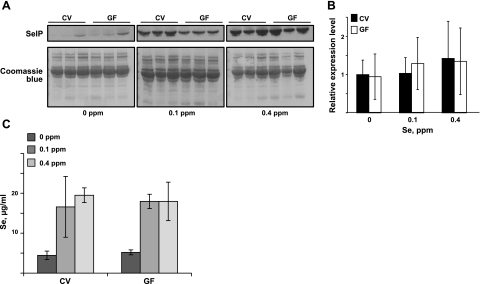
Mouse SelP contains 10 selenocysteine residues and is highly expressed in hepatocytes. This secreted protein transports Se from liver to other organs and was also suggested to serve an antioxidant function (22). Unlike other selenoproteins, SelP showed a trend toward increased levels in CV mice, at least in the mice fed 0.1 ppm Se, as examined by Western blots (Fig. 4A). However, its mRNA expression in the liver did not differ between GF and CV mice (Fig. 4B). We also examined Se levels in plasma in GF and CV mice (Fig. 4C). Whereas the 0-ppm Se group showed severalfold lower Se levels, there were no statistically significant differences within each dietary group between GF and CV mice.
Figure 4.
Regulation of SelP expression by dietary Se in CV and GF mice. A) SelP levels in plasma of CV and GF mice fed 0-, 0.1-, and 0.4-ppm Se diets, as analyzed by Western blots. B) Liver SelP mRNA expression analyzed by quantitative PCR. SelP mRNA expression in CV mice fed the 0-ppm Se diet was set to 1, and mRNA levels in other groups were calculated relative to this basal level. Significance was analyzed with Student's t test. C) Se levels in plasma in CV and GF mice fed 0-, 0.1-, and 0.4-ppm Se diets analyzed by ICP-MS. Values are expressed as means ± sd.
Influence of Se status and gut colonization on other trace elements
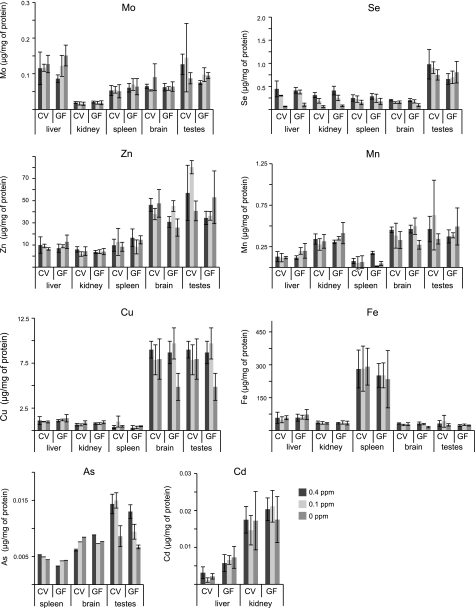
The intestinal microbiota may be sensitive to changes in trace element levels, and, in turn, gut colonization may influence host trace element status, either through competition for certain elements or by modifying the processes of food absorption or digestion. Using ICP-MS, we analyzed concentrations of several trace elements in CV and GF mice subjected to the 3 Se diets in liver, kidney, spleen, brain, and testes (Fig. 5). Se levels in liver, kidney, and spleen were sensitive to dietary Se, whereas its concentration in brain and testes did not differ with changes in the Se dietary levels. Also, we found no difference in the Se levels between GF and CV mice. Analysis of other trace elements, including Mn, Fe, Zn, Mo, Cu, and As, did not reveal statistically significant differences between Se diets. Cd levels in the liver were higher in GF mice regardless of the diet, suggesting a possible role of the gastrointestinal microbiota in assisting in detoxification of this element.
Figure 5.
Trace elements in CV and GF mice maintained on Se diets. Trace elements were analyzed with ICP-MS in tissues from CV and GF mice fed 0-, 0.1-, and 0.4-ppm Se diets, as described in Materials and Methods. Values are means ± sd. Organs in which trace elements were analyzed are indicated at the bottom of each panel.
DISCUSSION
Intestinal microbiota is sensitive to dietary Se status
Our results indicate that dietary Se can affect both the composition of the existing microbiota and establishment of gastrointestinal microflora. Particularly, in both experiments, the strongest effect was observed within genus Parabacteroides of the phylum Bacteriodetes, which demonstrated the opposite correlation with dietary Se supplementation. This finding might be explained by the use of Se by various microorganisms and Se toxicity to certain organisms. We also observed differential effects of Se across multiple phylotypes belonging to the phylum Bacteriodetes. It is possible that the niches vacated by the effect of Se on Parabacteroides were filled by other phylotypes within related taxonomic groups. The fact that multiple groups showed increases while fewer groups, in general, showed decreases is consistent with the overall effect of Se on increasing diversity of the microbiota. The differential sensitivity of various microorganisms to dietary Se is likely due to differences in their ability to uptake, store, use, and remove Se from the cell. In particular, the gut microbiota may sequester Se for expression of its own selenoproteins. About one quarter of all bacteria express selenoproteins, and, therefore, they require Se for optimal growth. For example, Escherichia coli has 3 selenoproteins. The number of selenoproteins in bacteria varies from 0 to 57. This use of Se by microorganisms decreases the availability of this trace element for the expression of host selenoproteins. As a result, microbiota increases the requirement of the host for Se. However, because of unavailability of the genomes for the majority of microorganisms that respond to Se status, it is not currently possible to directly link sensitivity to dietary Se and occurrence of specific Se utilization pathways.
Bacterial colonization decreases total Se status of the host
Se is an essential trace element required for synthesis and functions of 24 selenoproteins in mice. GPx1 and TR1 are among the best studied mammalian selenoproteins. TR1 is an essential selenoprotein that controls the redox state of thioredoxin. Thus, the thioredoxin system (which is the main redox regulatory system) is dependent on dietary Se. GPx1 is the most abundant mammalian selenoprotein that catalyzes glutathione-dependent reduction of hydroperoxides. It is expressed at particularly high levels in liver and kidney. MsrB1 is another important oxidoreductase, which repairs oxidatively damaged proteins by reducing methionine sulfoxide residues to methionines. Various selenoproteins are differentially regulated by dietary Se in mice. Some selenoproteins, such as GPx1 and MsrB1, are sensitive to Se status, whereas several other selenoproteins, including TR1, are less susceptible to changes in dietary Se. Thus, mice and other mammals have a priority for Se supply that is selenoprotein- and tissue-specific.
Under conditions of Se deficiency, GF mice were shown to possess higher GPx and TR activities in liver and intestine (16). They also had higher expression of GPx1 and its mRNA in the liver and colon. In addition, plasma, liver, and cecum of these mice contained higher Se concentration compared to CV animals (16). In our study, which compared GF mice with their CV controls, we also found GF mice to have elevated selenoprotein status, including higher GPx1 and MsrB1 activities and expression in liver and kidney. On the other hand, Se levels were similar in GF and CV mice subjected to the same Se diets. This observation might be explained by the fact, that in a previous study (16), GF and CV mice were subjected to the diets for 5 wk, whereas in our work, we conventionalized the GF mice with the intestinal flora prepared from C57BL/6J mice. Perhaps, at different Se levels in the diet, somewhat different populations of microorganisms colonize the gut.
In contrast to GPx1 and MsrB1 status in GF and CV mice, we observed a trend toward an increased level of SelP in CV mice. A previous study investigated gene expression in colonic epithelial cells, derived from germ-free and bacteria-reconstituted mice (23). SelP gene expression was found to be down-regulated under these conditions. Moreover, SelP was only detected in the GF mice. Since SelP is a transport form of Se, its expression contributes to the Se status of the whole organism. This protein is regulated by the glucocorticoid receptor (24) and by cytokines during inflammation. Its expression is decreased during certain pathologies linked to chronic inflammation, such as liver cirrhosis and Crohn's disease (22). In addition, it is induced by IL-10, an anti-inflammatory cytokine (24), but suppressed by IFN-γ, TNF-α, IL-1β, and TGF-β (25–27), which promote inflammation. The majority of plasma SelP comes from the liver. Its increase in CV mice might be explained by the indirect effect of cytokines on the expression of liver SelP or its increased use in organs of GF mice that are dependent on this protein for supplies of Se.
Microbiota and the host Se status
Disruption of integrity of the intestinal microbiota may lead to diseases, such as inflammatory bowel disease and cancer. Se supplementation may be beneficial in the prevention of colon cancer and certain other cancer types. Our data suggest that the sensitivity of the microbiota to dietary Se may be relevant to the regulation of the host Se status. Our diets were chosen to mimic the dietary Se intake in the human population. For example, the 0.1-ppm Se diet approximately corresponds to the human recommended dietary allowance for adults, whereas 0.4 ppm Se (17) may correspond to the diet supplemented with 200 μg Se/d, which is the dose most often used in human clinical trials involving Se. Thus, the effects observed with these diets in mice may be relevant to the effects of the human microbiota in setting the status of trace elements. In our experiments, the CV mice had higher requirements for Se compared to the GF mice, likely due to sequestration of Se by gut microorganisms, which compete for Se with the host. These data may also be relevant for understanding the mechanisms of Se-mediated protection against malignant transformation of the colon epithelium, but further studies are required to test this idea.
Acknowledgments
This study was supported by U.S. National Institutes of Health (NIH) grants GM061603 and AG021518 (to V.N.G.) and the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH (to D.L.H.).
REFERENCES
- 1. Possemiers S., Grootaert C., Vermeiren J., Gross G., Marzorati M., Verstraete W., Van de Wiele T. (2009) The intestinal environment in health and disease—recent insights on the potential of intestinal bacteria to influence human health. Curr. Pharm. Des. 15, 2051–2065 [DOI] [PubMed] [Google Scholar]
- 2. Hattori M., Taylor T. D. (2009) The human intestinal microbiome: A new frontier of human biology. DNA Res. 16, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fava F., Lovegrove J. A., Gitau R., Jackson K. G., Tuohy K. M. (2006) The gut microbiota and lipid metabolism: Implications for human health and coronary heart disease. Curr. Med. Chem. 13, 3005–3021 [DOI] [PubMed] [Google Scholar]
- 4. Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006) Microbial ecology: Human gut microbes associated with obesity. Nature 444, 1022–1023 [DOI] [PubMed] [Google Scholar]
- 5. Takaishi H., Matsuki T., Nakazawa A., Takada T., Kado S., Asahara T., Kamada N., Sakuraba A., Yajima T., Higuchi H., Inoue N., Ogata H., Iwao Y., Nomoto K., Tanaka R., Hibi T. (2008) Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int. J. Med. Microbiol. 298, 463–472 [DOI] [PubMed] [Google Scholar]
- 6. Tannock G. W. (2008) The search for disease-associated compositional shifts in bowel bacterial communities of humans. Trends Microbiol. 16, 488–495 [DOI] [PubMed] [Google Scholar]
- 7. Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 [DOI] [PubMed] [Google Scholar]
- 8. Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C., Hu C., Wong F. S., Szot G. L., Bluestone J. A., Gordon J. I., Chervonsky A. V. (2008) Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flint H. J., Duncan S. H., Scott K. P., Louis P. (2007) Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ. Microbiol. 9, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 10. Gibson G. R., Roberfroid M. B. (1995) Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 125, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 11. Baesman S. M., Bullen T. D., Dewald J., Zhang D., Curran S., Islam F. S., Beveridge T. J., Oremland R. S. (2007) Formation of tellurium nanocrystals during anaerobic growth of bacteria that use the oxyanions as respiratory electron acceptors. Appl. Environ. Microbiol. 73, 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stolz J. F., Basu P., Santini J. M., Oremland R. S. (2006) Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60, 107–130 [DOI] [PubMed] [Google Scholar]
- 13. Tompkins G. R., O'Dell N. L., Bryson I. T., Pennington C. B. (2001) The effects of dietary ferric iron and iron deprivation on the bacterial composition of the mouse intestine. Curr. Microbiol. 43, 38–42 [DOI] [PubMed] [Google Scholar]
- 14. Schomburg L., Schweizer U. (2009) Hierarchical regulation of selenoprotein expression and sex-specific effects of selenium. Biochim. Biophys. Acta 1790, 1453–1462 [DOI] [PubMed] [Google Scholar]
- 15. Irons R., Carlson B. A., Hatfield D. L., Davis C. D. (2006) Both selenoproteins and low molecular weight selenocompounds reduce colon cancer risk in mice with genetically impaired selenoprotein expression. J. Nutr. 136, 1311–1317 [DOI] [PubMed] [Google Scholar]
- 16. Hrdina J., Banning A., Kipp A., Loh G., Blaut M., Brigelius-Flohe R. (2009) The gastrointestinal microbiota affects the selenium status and selenoprotein expression in mice. J. Nutr. Biochem. 20, 638–648 [DOI] [PubMed] [Google Scholar]
- 17. Novoselov S. V., Calvisi D. F., Labunskyy V. M., Factor V. M., Carlson B. A., Fomenko D. E., Moustafa M. E., Hatfield D. L., Gladyshev V. N. (2005) Selenoprotein deficiency and high levels of selenium compounds can effectively inhibit hepatocarcinogenesis in transgenic mice. Oncogene 24, 8003–8011 [DOI] [PubMed] [Google Scholar]
- 18. Oh P. L., Benson A. K., Peterson D. A., Patil P. B., Moriyama E. N., Roos S., Walter J. (2010) Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 4, 377–387 [DOI] [PubMed] [Google Scholar]
- 19. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007) Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffmann P. R., Hoge S. C., Li P. A., Hoffmann F. W., Hashimoto A. C., Berry M. J. (2007) The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 35, 3963–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Novoselov S. V., Kim H. Y., Hua D., Lee B. C., Astle C. M., Harrison D. E., Friguet B., Moustafa M. E., Carlson B. A., Hatfield D. L., Gladyshev V. N. (2010) Regulation of selenoproteins and methionine sulfoxide reductases A and B1 by age, calorie restriction, and dietary selenium in mice. Antioxid. Redox Signal. 12, 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burk R. F., Hill K. E. (2009) Selenoprotein P-expression, functions, and roles in mammals. Biochim. Biophys. Acta 1790, 1441–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fukushima K., Ogawa H., Takahashi K., Naito H., Funayama Y., Kitayama T., Yonezawa H., Sasaki I. (2003) Non-pathogenic bacteria modulate colonic epithelial gene expression in germ-free mice. Scan. J. Gastroenterol. 38, 626–634 [DOI] [PubMed] [Google Scholar]
- 24. Rock C., Moos P. J. (2009) Selenoprotein P regulation by the glucocorticoid receptor. Biometals 22, 995–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bosschaerts T., Guilliams M., Noel W., Herin M., Burk R. F., Hill K. E., Brys L., Raes G., Ghassabeh G. H., De Baetselier P., Beschin A. (2008) Alternatively activated myeloid cells limit pathogenicity associated with African trypanosomiasis through the IL-10 inducible gene selenoprotein P. J. Immunol. 180, 6168–6175 [DOI] [PubMed] [Google Scholar]
- 26. Mostert V., Dreher I., Kohrle J., Wolff S., Abel J. (2001) Modulation of selenoprotein P expression by TGF-beta(1) is mediated by smad proteins. BioFactors 14, 135–142 [DOI] [PubMed] [Google Scholar]
- 27. Yi Y. S., Park S. G., Byeon S. M., Kwon Y. G., Jung G. (2003) Hepatitis B virus X protein induces TNF-alpha expression via down-regulation of selenoprotein P in human hepatoma cell line, HepG2. Biochim. Biophys. Acta 1638, 249–256 [DOI] [PubMed] [Google Scholar]