Abstract
Restriction endonucleases were tested for their ability to catalyze the cleavage of mismatch-containing recognition sites in DNA. These mismatched base pairs were T.G, U.G, or A.C in covalently closed, circular heteroduplexes prepared by in vitro extension of chemically synthesized oligonucleotide primers annealed to a bacteriophage M13-derived viral DNA. None of the restriction enzymes was able to completely cleave the mismatch-containing recognition sites under standard conditions. However, three of them, SmaI, SalI, and SstI, catalyzed partial digestion leading to an accumulation of DNA singly nicked at the mismatched recognition site. The ability of SmaI and SstI to partially cleave at a mismatch was shown to depend on the nature and position of the mismatch within the corresponding recognition site. In contrast, little or no digestion was obtained with AccI, HincII, HindIII, and KpnI at mismatch-containing sites. Therefore, in some cases a transition-type substitution in only one strand of a recognition site inhibits restriction endonuclease-catalyzed digestion at that site although in others partial digestion occurs.
Full text
PDF



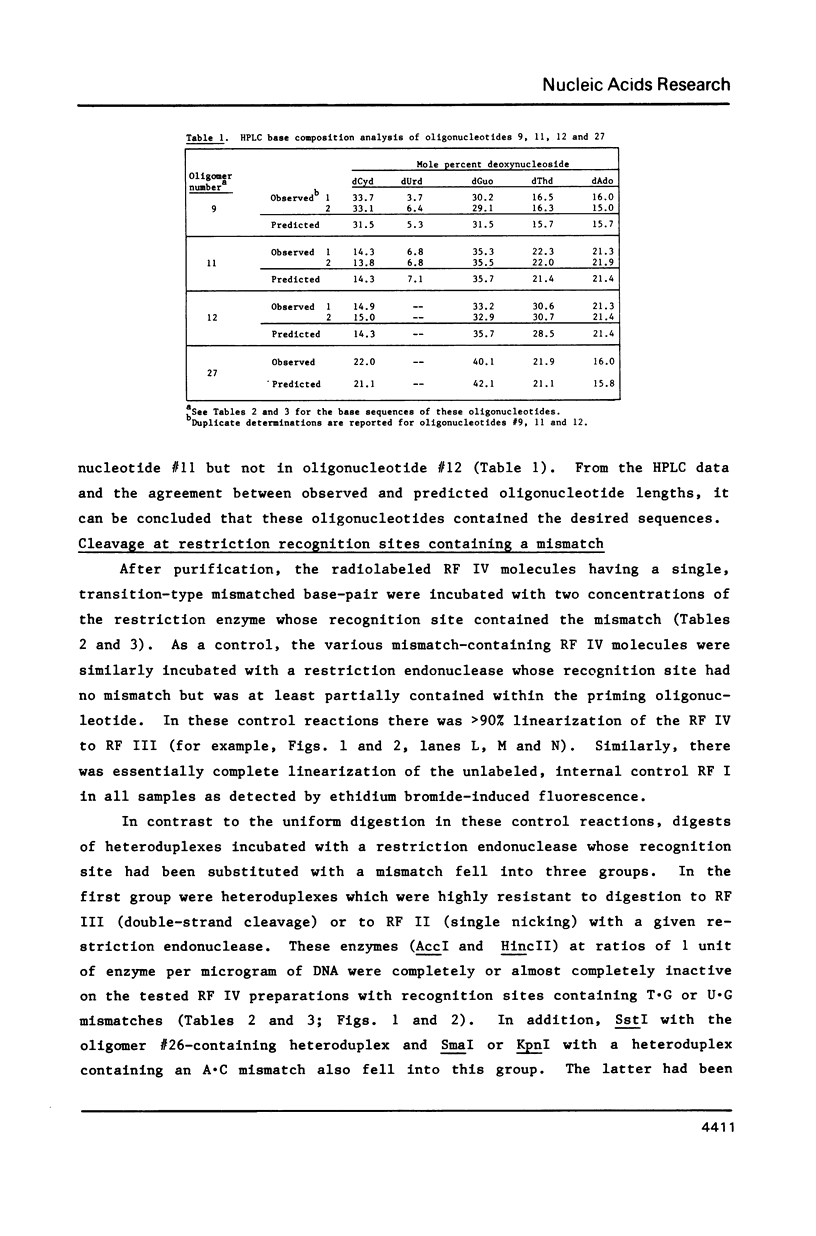
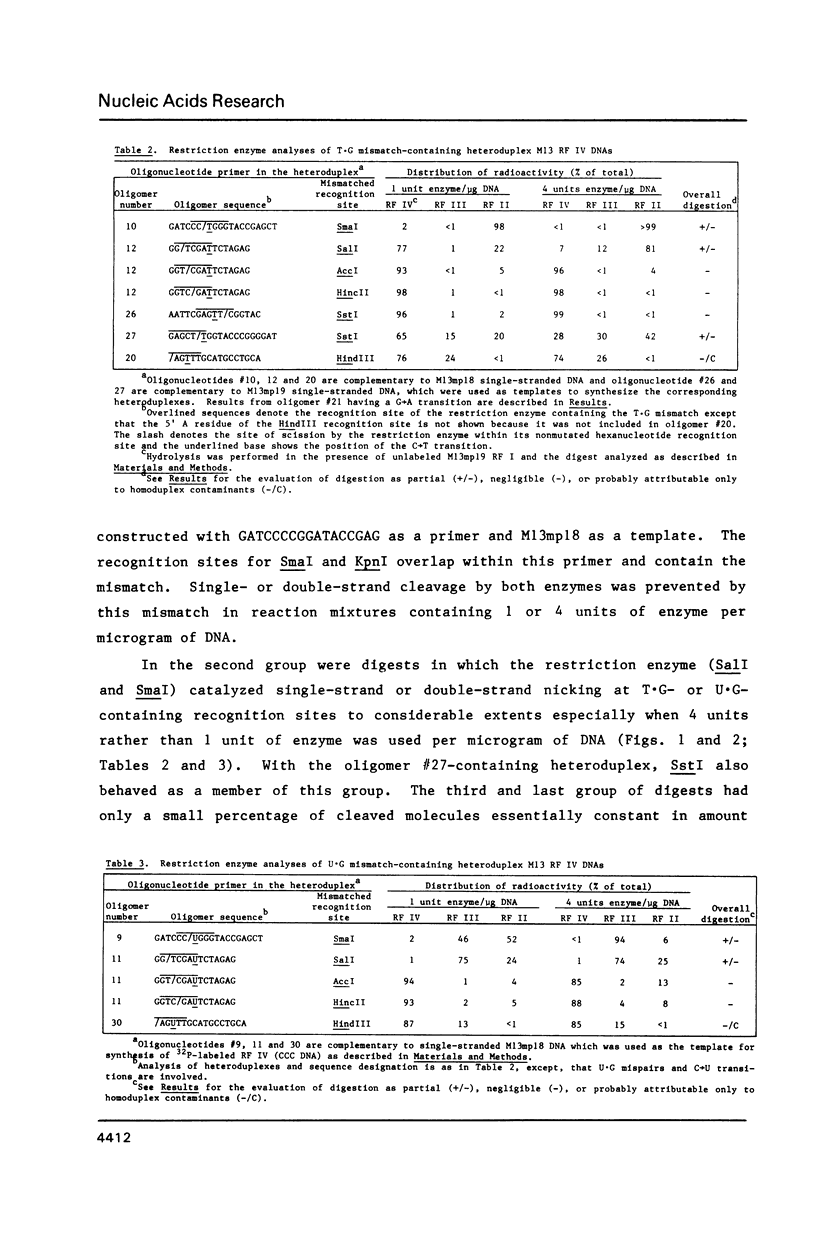








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arraj J. A., Marinus M. G. Phenotypic reversal in dam mutants of Escherichia coli K-12 by a recombinant plasmid containing the dam+ gene. J Bacteriol. 1983 Jan;153(1):562–565. doi: 10.1128/jb.153.1.562-565.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark B. R., Roberts R. J., RajBhandary U. L. Use of short synthetic DNA duplexes as substrates for the restriction endonucleases Hpa II and Mno I. J Biol Chem. 1979 Sep 25;254(18):8943–8950. [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. The effects of substituted pyrimidines in DNAs on cleavage by sequence-specific endonucleases. J Biol Chem. 1979 Apr 10;254(7):2551–2560. [PubMed] [Google Scholar]
- Bodnar J. W., Zempsky W., Warder D., Bergson C., Ward D. C. Effect of nucleotide analogs on the cleavage of DNA by the restriction enzymes AluI, DdeI, HinfI, RsaI, and TaqI. J Biol Chem. 1983 Dec 25;258(24):15206–15213. [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P., Poltev V. I. Alteration of the DNA double helix conformation upon incorporation of mispairs as revealed by energy computations and pathways of point mutations. Nucleic Acids Res. 1985 Jan 11;13(1):141–154. doi: 10.1093/nar/13.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R., Blakesley R. Guide to the use of type II restriction endonucleases. Methods Enzymol. 1983;100:3–38. doi: 10.1016/0076-6879(83)00043-9. [DOI] [PubMed] [Google Scholar]
- Gehrke C. W., McCune R. A., Gama-Sosa M. A., Ehrlich M., Kuo K. C. Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J Chromatogr. 1984 Sep 28;301(1):199–219. doi: 10.1016/s0021-9673(01)89189-5. [DOI] [PubMed] [Google Scholar]
- Gillam S., Smith M. Site-specific mutagenesis using synthetic oligodeoxyribonucleotide primers: I. Optimum conditions and minimum ologodeoxyribonucleotide length. Gene. 1979 Dec;8(1):81–97. doi: 10.1016/0378-1119(79)90009-x. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981 Jun 11;9(11):2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P. Single turnovers of the EcoRI restriction endonuclease. Biochem J. 1983 May 1;211(2):405–415. doi: 10.1042/bj2110405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. H., Farnet C. M., Ehrlich K. C., Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982 Mar 11;10(5):1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J., Martin D. Restriction endonucleases HindII and TaqI cleave DNA with mismatched nucleotides within their recognition sequences. Nucleic Acids Res. 1986 Mar 11;14(5):1943–1949. doi: 10.1093/nar/14.5.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. A., Nierlich D. P. Cleavage of Nonglucosylated Bacteriophage T4 deoxyribonucleic acid by Restriction Endonuclease Eco RI. J Biol Chem. 1975 Mar 25;250(6):2395–2397. [PubMed] [Google Scholar]
- Klenow H., Overgaard-Hansen K., Patkar S. A. Proteolytic cleavage fo native DNA polymerase into two different catalytic fragments. Influence of assay condtions on the change of exonuclease activity and polymerase activity accompanying cleavage. Eur J Biochem. 1971 Oct 14;22(3):371–381. doi: 10.1111/j.1432-1033.1971.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Landy A., Ruedisueli E., Robinson L., Foeller C., Ross W. Digestion of deoxyribonucleic acids from bacteriophage T7, lambda, and phi 80h with site-specific nucleases from Hemophilus influenzae strain Rc and strain Rd. Biochemistry. 1974 May 7;13(10):2134–2142. doi: 10.1021/bi00707a022. [DOI] [PubMed] [Google Scholar]
- Marchionni M. A., Roufa R. J. Digestion of 5-bromodeoxyuridine-substituted lambda-DNA by restriction endonucleases. J Biol Chem. 1978 Dec 25;253(24):9075–9081. [PubMed] [Google Scholar]
- Maxwell A., Halford S. E. The SalGI restriction endonuclease. Mechanism of DNA cleavage. Biochem J. 1982 Apr 1;203(1):85–92. doi: 10.1042/bj2030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Modrich P., Rubin R. A. Role of the 2-amino group of deoxyguanosine in sequence recognition by EcoRI restriction and modification enzymes. J Biol Chem. 1977 Oct 25;252(20):7273–7278. [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1984;12 (Suppl):r167–r204. doi: 10.1093/nar/12.suppl.r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S., Ehrlich M. Isolation of in vitro synthesized covalently closed circular double-stranded DNA by selective denaturation and filtration. Prep Biochem. 1984;14(5):485–497. doi: 10.1080/00327488408061782. [DOI] [PubMed] [Google Scholar]
- Smith H. O. Nucleotide sequence specificity of restriction endonucleases. Science. 1979 Aug 3;205(4405):455–462. doi: 10.1126/science.377492. [DOI] [PubMed] [Google Scholar]
- Streeck R. E. Single-strand and double-strand cleavage at half-modified and fully modified recognition sites for the restriction nucleases Sau3a and Taqi. Gene. 1980 Dec;12(3-4):267–275. doi: 10.1016/0378-1119(80)90109-2. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Shedlarski J. G., Farber M. B., Kuebbing D., Ehrlich M. Two sequence-specific endonucleases from Xanthomonas oryzae. Characterization and unusual properties. Biochim Biophys Acta. 1980 Feb 29;606(2):371–385. doi: 10.1016/0005-2787(80)90047-7. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Shenoy S., Ehrlich M. DNA methylation inhibits the transfecting activity of replicative- form phi X174 DNA. J Virol. 1984 Mar;49(3):674–679. doi: 10.1128/jvi.49.3.674-679.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder N. D., Boeke J. D. The filamentous phage (Ff) as vectors for recombinant DNA--a review. Gene. 1982 Jul-Aug;19(1):1–10. doi: 10.1016/0378-1119(82)90183-4. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]