Abstract
Genetic factors contribute upto 15%–30% cases of male infertility. Formation of spermatozoa occurs in a sequential manner with mitotic, meiotic, and postmeiotic differentiation phases each of which is controlled by an intricate genetic program. Genes control a variety of physiologic processes, such as hypothalamus–pituitary–gonadal axis, germ cell development, and differentiation. In the era of assisted reproduction technology, it is important to understand the genetic basis of infertility to provide maximum adapted therapeutics and counseling to the couple.
Keywords: Azoospermia, cytogenetic, DNA damage, epigenetics, infertility, mtDNA, mRNA, oligozoospermia, Y chromosome
INTRODUCTION
There is increasing recognition of the contribution of genetic abnormalities to the causation of male infertility. The genetic factors involved in male infertility may be chromosomal or monogenic disorders, mitochondrial DNA (mtDNA) mutations, Y chromosome deletions, multifactorial disorders, imprinting disorders, or endocrine disorders of genetic origin. About 10% genes in the genome are related to spermatogenesis. Genetic abnormalities account for 15%–30% cases of male infertility[1] and may lead to irreversible partial or complete spermatogenic arrest. With the increased use of assisted reproduction technology (ART), our understanding of genetic basis of male infertility has large implications not only for understanding the cause of infertility but also in determining the prognosis and management of such couples.
CHROMOSOMAL ABNORMALITIES ASSOCIATED WITH INFERTILITY
Chromosomal abnormalities include both numerical and structural chromosomal aberrations, which are large enough (>4–5 Mb) to be discerned under the microscope. They account for 5%–10% cases of oligozoospermia, to 15%–25% cases with nonobstructive azoospermia.[1–3] In azoospermia, sex chromosome abnormalities are predominant. However, autosomal structural chromosomal abnormalities are the chief genetic cause of oligozoospermia and may be balanced (no gain or loss of genetic material) or unbalanced.
NUMERICAL ABERRATIONS
Numerical chromosome errors are a frequent cause of male infertility. The incidence is inversely proportional to the number of sperm in ejaculate.
47, XXY
Klinefelter syndrome [KFS] is the most common numerical chromosomal abnormality and is the most common cause of azoospermia. It is found in 11% cases with azoospermia. Men with 47,XXY [Figure 1] chromosomal complement are azoospermic due to seminiferous tubule dysgenesis, whereas mosaic cases with normal 46,XY cell line may be oligozoospermic. Identification of normal cell line is of critical importance as it indicates the presence of isolated foci of spermatogenesis, and thus is a good prognostic factor for ART. Studies on sperm chromosome from Klinefelter men have shown that the extra chromosome is eliminated during spermatogenesis.[4] Although majority of offspring from such men have normal chromosomal complement, the risk of chromosomally abnormal fetus is also high.[5] Frequency of aneuploidy for the sex chromosomes varies from 1.5%[6] to 7%[7] in sperm from Klinefelter mosaics, and 2%[8] to 45%[9] in the sperm of men who appear to have a nonmosaic 47,XXY karyotype.
Figure 1.
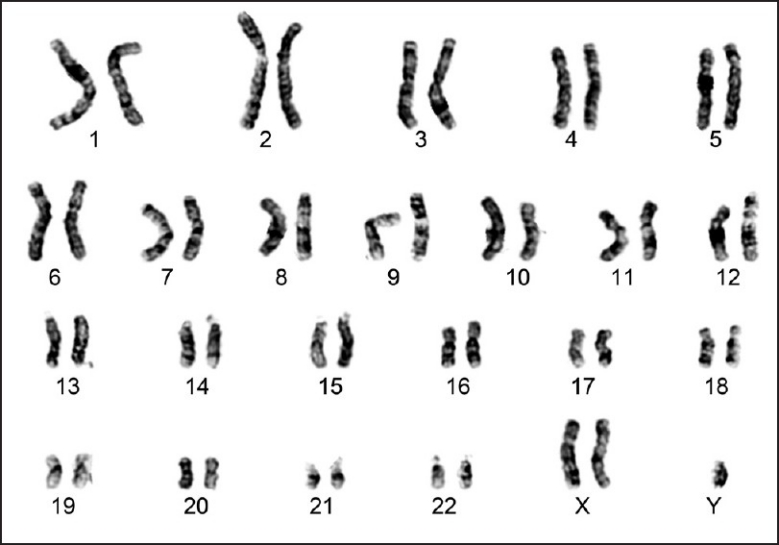
G-banded karyotype showing 47,XXY chromosomal complement (Klinefelter syndrome)
46,XX male
This disorder affects 1 in 20,000 men. During paternal meiosis, aberrant translocation of Y material, including sex determining region (SRY) to the X chromosome leads to the 46,XX male chromosomal complement. Presence of SRY gene results in testicular differentiation but absence of spermatogenesis due to absence of long arm of Y chromosome.[10] Such men have normal sexual development with normal external genitilia but increased incidence of hypospadias and cryptorchidism.
STRUCTURAL ABERRATIONS
Translocations
Robertsonian translocations
Robertsonian translocations (RT) are found in 1 in 1000 individuals[11] and are the most common structural rearrangements in infertile men.[12] RT result in the fusion of the long arm of 2 acrocentric (Group D chromosomes: 13, 14, 15 and Group G chromosomes: 21, 22, and Y) chromosomes [Figure 2]. The fused short arms are mostly lost, and hence the carrier has a chromosomal constitution with 45 chromosomes. When the chromosomes pair during meiosis, they do so as a trivalent, and the resulting gametes can be chromosomally normal or aneuploid with an extra or missing long arm of chromosome.
Figure 2.
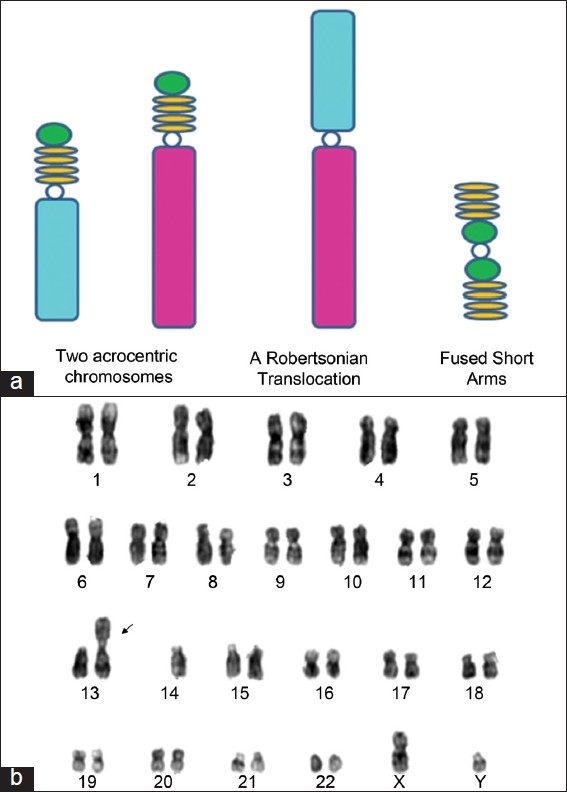
(a) Schematic illustration of Robertsonian translocation; (b) G-banded karyotype showing 45,XY t (13;14) chromosomal complement
Reciprocal translocations
Reciprocal translocations occur when there is exchange of genetic material between nonhomologous chromosomes. It affects 1.17% of infertile men.[13] During meiosis, 4 chromosomes pair as quadravalents and their segregation results in a higher frequency of unbalanced chromosomes as compared with carriers of RT.
Risk of meiotic imbalance is primarily determined by the characteristics of the chromosomes involved, and the breakpoint positions. Many of these imbalances leads to fetal mortality and the average frequency of paternally derived translocation imbalance at prenatal diagnosis is 12%.[14] The frequency of chromosome abnormality is high, so these couples should undergo preimplantation genetic diagnosis (PGD) to implant only chromosomally normal or balanced embryos.
Inversions
These balanced structural rearrangements are found in 10%–15%[15] of prenatal diagnosis and 0.1% of infertile men.[13] Inversion occurs when 2 chromosome breaks occur on the same chromosome and join after 180° rotation. Inversion may be
Paracentric inversions
Paracentric inversions occur when both breakpoints are on one chromosome arm and do not involve the centromere.
Pericentric inversions
Pericentric inversions occur when the chromosome breaks include the centromere and the short and long arm of the chromosome.
The inversions that produce recombinant chromosomes are, in general, large inversions encompassing more than half of the chromosome length.
Germline mosaics
Germline mosaics are men with a normal somatic karyotype but an abnormal cell line in their germ cells. They are discovered only on a testicular biopsy. Studies have discovered that 1%–17% of infertile men have germline mosaicism.[16,17] The increased frequency of chromosomal abnormalities in intracytoplasmic sperm injection pregnancies and newborns reflects the increased frequency observed in the sperm of infertile men. Furthermore, studies have indicated that these chromosomal abnormalities are of paternal origin,[18] highlighting the fact that chromosomally abnormal sperm in such patients may lead to chromosomally abnormal offspring.
It has also been suggested that abnormalities of the centrosome may exist in surgically retrieved sperm, a problem that may lead to increased mitotic nondisjunction and mosaicism in resulting embryos.[19]
GENES ASSOCIATED WITH INFERTILITY
Y chromosome genes
Y chromosome [Figure 3] is one of the smallest human chromosomes (60 Mb) and is highly polymorphic in length. It is male specific and the only haploid component of the human genome. Till now 156 transcription units, 78 protein coding genes, and 27 distinct proteins (9 on Yp and 18 on Yq) encoded by the Y chromosome have been identified.
Figure 3.
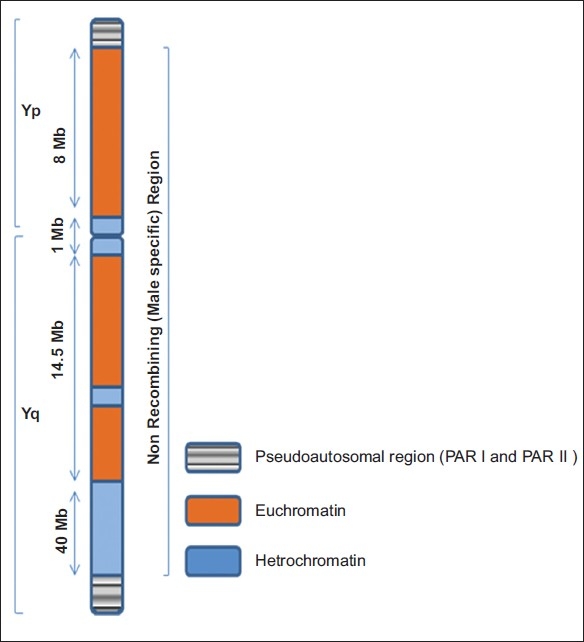
Schematic representation showing human Y chromosome showing pseudoautosomal (PAR I and PAR II) and nonrecombining regions (MSY)
The Y chromosome is divided into 7 deletions intervals. The region critical for spermatogenesis is on interval 5 and 6. The length of euchromatic DNA sequences on the Y is about 23 Mb, including 8 Mb on the short arm and 14.5 Mb on the long arm. There are 3 classes of euchromatic sequences those transposed from the X chromosome during the process of the evolution of the Y (X-transposed), those similar to sequence information from the X chromosome (X-degenerate), and those repeated units across the proximal short arm of the Yp and across most of the Yq (amplicons). Within the X transposed segments, only 2 protein-encoding genes have been identified (TGIF2LY and PCDH11Y). The X-degenerate regions, with a combined length of 8.5 Mb, are dotted with single-copy genes or pseudogenes that are mostly expressed ubiquitously (ie, expressed in multiple organs in the body and not confined to a specific tissue). The sex-determining gene (SRY) is located in this region. The SRY gene expresses a transcription factor that switches on the genes for male sexual differentiation.[20] The genes in the AZFa (DBY and USP9Y) are located in the X-degenerate region. The most complex regions of the Y chromosome are the unique ampliconic regions in the euchromatin that are 10.5 Mb in length.[20,21]
Amplicons are families of units composed of nucleotide sequences that are similar to each other. They are located in 7 segments that are scattered across the euchromatin on Yp and Yq. The amplicons harbor the highest density of the Y chromosome genes that are exclusively expressed in the testes and enhance sexual fitness. Genes related to the AZFb and AZFc are located in the ampliconic regions. The array of the amplicons forms 8 palindromes (P1–P8).[20] A palindrome is a DNA sequence containing different amplicons, which has a twin along the chromosome that read the same in a reverse direction. Most of the recognized genes that are deleted in infertile men are located in the palindromic regions of the Yq.
Microdeletion of the Y chromosome
Microdeletions on the long arm of the Y chromosome (Yq) is one of the most significant pathogenic defects associated with male infertility. Tiepolo and Zuffardi in 1976, hypothesized a correlation between Y chromosome deletions and male infertility.[22] Yq microdeletions are found in 13% azoospermic men, 1%-7% severely oligozoospermic men (sperm count less than 0.5 million/mL). These deletions are clustered in interval 5 and 6 of the Y chromosome. It was defined as Azoospermia Factor (AZF) as the most severe phenotype associated with its deletion was azoospermia. The AZF region is further subdivided into 3 nonoverlapping regions termed as AZFa, AZFb, and AZFc. Six genes located in the AZF regions are expressed exclusively in the testes and are therefore known as “AZF candidate genes.”
The AZFa region
The AZFa spans around 400-600 kb of DNA and is located in the proximal portion of deletion interval 5. AZFa region harbors 2 protein encoding genes, namely, USP9Y and DBY (recently termed DDX3Y).
Deletions of AZFa loci is characterized by Sertoli-cell-only syndrome, type I.[23]
The AZFb region
The AZFb spans around 1–3 Mb of DNA and is located on the distal portion of deletion interval 5 to the proximal end of deletion interval 6 (subinterval 5O–6B). Protein-encoding genes in AZFb region are EIF1AY, RPS4Y2, and SMCY that are located in X-degenerate euchromatin, and HSFY, XKRY, PRY, and RBMY that are in the ampliconic region.[23] The RBMY gene family encodes testis-specific RNA-binding proteins that are involved in mRNA processing, transport, and splicing. These are exclusively expressed in the germ cells and are 6 copies of this gene family in the AZFb region. AZFb gene is expressed in primary spermatocytes and thus its deletion leads to the arrest of meiosis and manifests as maturation arrest with the accumulation of primary spermatocytes.
The AZFc region
AZFc spans 3.5 Mb of euchromatin and is located at the distal part of deletion interval 6 (subinterval 6C-6E) on the Y chromosome.[22] Deletions of the AZFc region are most common in men with idiopathic oligozoospermia or azoospermia. In a study in our laboratory we found that 14% of the cases harbored AZFc deletion.[2] The AZFc region contains 8 gene families that are involved in spermatogenesis—BPY2, CDY, DAZ, CSPG4LY, GOLGAZLY, TTY3.1, TTY4.1, and TTY7. There are 4 copies of DAZ and it also has an autosomal homolog DAZL on 3p24. The first recognized gene in the AZFc was Deletion in Azoospermia factor (DAZ). This gene encodes RNA-binding proteins that are exclusively expressed in the germ cells.[20] Deletion of each member of DAZ may have different effects. Deletions in DAZ2, DAZ3, and DAZ4 copies are found in both fertile and infertile men and are described as familial variants inherited from father to son. However, DAZ1/ DAZ2 deletions are reported to be restricted only to infertile men. Deletions of AZFc regions are associated with a variable phenotype ranging from hypospermatogenesis to maturation arrest. Cases with AZFc deletions show a progressive deterioration in spermatogenesis and cases develop azoospermia over a period of time.[24]
gr/gr deletions
A number of infertile men harbor partial deletions in AZFb and AZFc loci. Considering the Y chromosome structure and the complexity of the deletion mechanism involved, it is possible that few other deletions also supplement the effect of partial AZFb and AZFc deletions. Frequency and pathologic significance of these partial deletions is still not known. de Llanos et al. reported a partial 1.6 Mb deletion on Y chromosome, termed as “gr/gr” deletion specifically in infertile men with varying degrees of spermatogenic failure.[25]
The gr/gr deletion removes part of the AZFc region, including 2 copies of DAZ, 1 copy of CDY1, and a few transcription units. Since father-to-son transmission is observed, the gr/gr deletion probably results in subfertility rather than infertility.
Another deletion termed as b2/b3 or u3-gr/gr or g1/g3, which removes a similar quantity of AZFc genes, has also been identified.[26]
CLINICAL IMPLICATIONS OF Y CHROMOSOME AZF DELETION
AZF deletions are specific for spermatogenic failure. The deletion of AZFa or AZFb loci and large deletions encompassing 2 or more loci lead to azoospermia and carry poor prognosis if such couples opt for ART. Cases with AZFc deletions have a variable phenotype and chances of sperm retrieval are high; however, such cases show progressive decline in sperm count over time and progress from oligozoospermia to azoospermia. [24] Such men are good candidates for ART but chances of iatrogenic transmission of Yq microdeletion to offspring must be explained to the couple.
AUTOSOMAL GENE MUTATIONS IN INFERTILITY
The complex interaction of gene products on both the sex chromosomes and the autosomes are essential for the fertility of an individual. Several autosomal genes, such as acrosin gene, BAX, BCL16, c-kit, ATM, HSP70.2, RAD6B, MDHC7, CREM, and DNA11 and 12, play an important role in germ cell development and spermiogenesis. Mutations of CFTR gene on chromosome 7 leads to obstructive azoospermia and is found in 60%–90% of patients with congenital bilateral absence of vas deferens (CBAVD). F508 del is the most common CFTR mutation found in 60%–70% of CBAVD patients. If both the partners have CFTR mutation, PGD should be done to avoid cystic fibrosis in the offspring.[27]
Gene located on the chromosome 2 encodes for receptor of the follicle stimulating hormone and is another autosomal gene essential for fertility. Although there is no sufficient data correlating FSHR gene mutations and infertility in men, polymorphism of the FSHR gene affects the FSH receptivity in women. A few studies have correlated the FSHR gene mutations and abnormal spermatogenesis.[28]
The methylenetetrahydrofolate reductase (MTHFR) plays a critical role in folate metabolism, and is essential for DNA methylation and spermatogenesis. The gene for MTHFR is located on chromosome 1 and any mutation in the MTHFR may disrupt the methylation of the nucleotides in the germ cells, a very sensitive regulatory step for the accurate transmission of genetic information to the offspring. The valine to alanine substitution due to C667T polymorphism in this gene reduces the activity of the enzyme. This reduced activity may lead to dysregulation of folic acid metabolism, ultimately manifesting as inaccurate methylation of nucleotides and subsequently impairing spermatogenesis.[29]
Steroid 5-alpha reductase converts testosterone into dihydrotestosterone (DHT); a more active metabolite. DHT is required for spermatozoa maturation in the epididymis.[30] SRD5A1 and SRD5A2 are the 2 isozymes providing 5-alpha reductase activity.[31] SRD5A1 is expressed mainly in skin, whereas SRD5A2 in seminal vesicles.[32] Mutations in SRD5A2 gene located on chromosome 2, cause deficient virilization and pseudohermaphroditism.[33] Missense mutations in SRD5A2 gene affects the 5-alpha reductase activity. A valine-to-leucine and an alanine-to-threonine substitution at codon 89 and 49, respectively, has a higher frequency as compared with other amino acids, altering sequence variations.[34]
Inguinoscrotal descent is controlled by several genes, namely, INSL3, LGR8, HOXA10, HOXA11, and ARID5B. Approximately 5% of cryptorchoid men have mutations in these genes.[35]
PROTAMINE AND TRANSITIONAL PROTEIN GENES
Gradual replacement of histones by transitional proteins (TP) followed by protamines (PRM) play a crucial role in sperm nuclear condensation. Equal proportions of PRM1 and PRM2 are present in spermatozoa of normal men, whereas the PRM1/PRM2 ratio is altered in infertile men.[36,37] These altered levels could be either due to the pathogenic mutation in PRM/TNP genes or aberrant posttranslation modification of mRNAs. Thus abnormal condensation of sperm nucleus may lead to increased sperm DNA fragmentation, resulting in decreased sperm function, thus affecting male fertility.
X-Linked genes
Although it is believed that Y chromosome plays a major role in fertility regulation, several genes located on X chromosome also regulate fertility.
Listed below are a few genes and syndromes with infertility as one of the clinical features, which are associated with genes on the X chromosome.
Androgen insensitivity syndrome (AIS/IMS) is typically characterized by evidence of feminization of the external genitalia at birth, abnormal secondary sexual development at puberty, and infertility in individuals with a 46,XY karyotype.[38,39] AIS represents a spectrum of defects in androgen action and can be subdivided into 3 broad phenotypes: complete AIS, with typical female genitalia; partial AIS with predominantly female, predominantly male, or ambiguous genitalia; and mild AIS with typical male genitalia.[40–43] AIS is inherited in an X-linked recessive manner. Affected 46,XY individuals are almost always infertile. Carrier females have a 50% chance of transmitting the AR gene (Xq11-12) mutation in each pregnancy.
The USP26 gene located on the long arm of X chromosome is expressed in the preliminary stages of spermatogenesis. It is involved in histone removal during protamination. Increased histone levels in sperm DNA after complete differentiation is associated with increased sperm DNA damage.
TAF7L is an X-linked (Xq22.1) germ cell-specific paralog of TAF7, which is generally expressed as a component of TFIID. It is known from studies that TAF7L gene is expressed during spermatogenesis, from the stage of spermatogenesis till the stage of round spermatids.[44,45]
The gene for Kallmann syndrome (KS) (KAL1), a form of idiopathic hypogonadotropic hypogonadism, is on the short arm of X chromosome. Deletions of KAL1 were found in 30%-70% cases of KS. The gene for the X-linked form was mapped to Xp22.3, spans 200,000 bp genomic DNA. Many different mutations in this gene, variably termed as KAL-X, KALIG-1, or ADMLX, were described in individuals with KS.
SYNDROMES ASSOCIATED WITH MALE INFERTILITY
Important syndromic cases linked to male infertility are KFS, AIS/IMS, KS, Kartagener syndrome, metabolic syndrome, Persistent Mullerian duct syndrome, Aarskog–Scott syndrome, Kearns–Sayre syndrome, Polyglandular failure syndrome types I and II, Bardet–Biedl syndrome, Noonan syndrome, Prader–Willi syndrome, deafness–infertility syndrome,[46] and others, which are yet to be investigated. These syndromic features may provide an early clue to the physician for the better treatment of the patients.
Kartagener syndrome is a form of primary ciliary dyskinesia, which includes bronchiectasis, sinusitis, and sperm immotility with situs inversus.
Metabolic syndrome (MetS) represents a constellation of abnormalities, including dyslipidemia, hypertension, and impaired glucose metabolism, with insulin resistance as the hypothesized underlying pathogenic mechanism. Male factor infertility may represent one such perturbation in some male patients with MetS. MetS is an important medical and epidemiologic entity, as its deleterious effect on patients is firmly established. Male infertility may represent another aberration observed in some patients with MetS. Currently, there is sufficient evidence to suggest the MetS–male infertility paradigm.[47] Obesity/overweight may result in hypogonadism, increased scrotal temperatures, impaired spermatogenesis, decreased sperm concentration and motility, and increased sperm DNA damage. Similarly, non insulin dependent diabetes melitus (NIDDM)/insulin resistance may contribute to and compound this scenario. Dyslipidemia with increased oxidative stress in the testicular microenvironment and/or excurrent ductal system may further decrease fertility.
Kearns–Sayre syndrome belongs to a group of multisystemic disorders caused by mutations in the mitochondrial genome. This syndrome most prominently involves the neuromuscular and endocrine system. Pathology of the reproductive system is reported in 20%–30% affected males. Abnormal findings include cryptorchidism, pubertal delay, subnormal testicular volume, and low gonadotropin levels.
EPIGENETICS AND INFERTILITY
Difference in cells despite having the same genome arises because of the differential pattern of gene expression. Differential gene expression is produced by variability in the DNA-associated protein and its modifications (methylation, demethylation, acetylation, and deacetylation). These modifications are responsible for variable gene expression and constitute an integral component of epigenetics. Epigenetics has critical role in sperm development and function, fertilization, and postfertilization events.
In the postmeiotic stage of spermatogenesis, transition proteins (TNP1 and TNP2) replace histones, which in turn are replaced with protamines. During the histone to transition protein substitution, core histone tail is hyperacetylated. The acetylation pattern determines the transcriptional status of a gene, hyperacetylation marking the gene with high transcriptional activity. A slight anomaly of this process can lead to sperm dysgenesis or aberration in chromatin packaging in spermatozoa, leading to infertility.
Ratio between protamine1 (P1) and protamine2 (P2) acts as a marker of male fertility. In normal fertile men, this ratio (P1/P2) is 0.8 to 1.2.[48–50] Any deviation from this ratio is indicative of increased susceptibility to DNA damage and poor semen quality.[51–53] Phosphorylation and dephosphorylation of protamines also play a vital role in sperm function, for example, CamK4 phosphorylates protamine2 and it is observed that mutations in CamK4 lead to derangement in spermatogenesis, and therefore male infertility. Abnormalities in protamine packaging of DNA causes aberrant gene expression resulting in either hypertranscription or transcriptional arrest, leading to failure in spermatogenesis.[54] Studies suggest that spermatozoa convey a wide variety of epigenetic information to the oocyte, which has an important role in postfertilization events.
DNA in sperm is not homogeneously packed. The peripheral portion of nuclear DNA (15%) is packaged with histones and the inner core of DNA is a highly compact crystalline structure bound to protamines.[55] Ooplasm plays an important role in demethylation of paternal DNA after fertilization. The nucleosomes derived from spermatozoa remain attached to paternal genome in zygote before the S-phase,[56] which gives a clue that these nucleosomes might be involved in epigenetic functions in postfertilization developmental events.
Brother of regulator of imprinting sites (BORIS), an epigenetic factor specifically expressed in male gonads plays a crucial part in methylation levels in male germ cell differentiation.[57] BORIS along with other factors (CCCTC-binding factor and insulator protein) binds to specific DNA regions (during spermatogenesis) and prevent interaction between promoter and enhancer region, which as a consequence modulates spermatogenesis-specific gene expression, producing normal sperms. Thus BORIS plays a decisive role in spermatogenesis and male fertility.[58]
MITOCHONDRIAL DNA MUTATIONS AND INFERTILITY
mtDNA is circular DNA molecule that codes for 13 polypeptides of the oxidative phosphorylation pathway. Mitochondrial DNA is a naked molecule, lacks introns and histones and both strands are transcribed to synthesize a functional protein. mtDNA mutates at a rate 10–20 times higher than nuclear DNA due to asexual method of replication, a very basic repair mechanism, lack of protective histones, and increased proximity to the site of free radical generation. Mitochondria are located in sperm midpiece and each mitochondria has 1 mtDNA copy.[59–61]
Spermatozoa exist in a state of oxygen paradox and have higher energy requirement to support their movement after ejaculation. Thus, random attacks on the naked mtDNA of sperms by reactive oxygen species (ROS) or free radicals causes oxidative damage or mutation to the mt genome, which leads to infertility.[62,63]
In infertile men, 3 large deletions occur in higher frequency as compared with fertile men. The 4977-bp, 7345-bp, and 7599-bp deletions in the sperm genome are detrimental to the sperm reproductive potential because these deletions span the ATPase 8 and 6, COII, ND3, ND4, ND5, ND6, and cytochrome b oxidase coding components of the oxidative phosphorylation (OXPHOS) pathway.[64,65]
A 15-bp deletion in the cytochrome c oxidase subunit (COX III) of complex IV is also reported to be associated with motility loss in sperms.[66] Two base pair deletions at 8195-8196 in the CO II gene and a A3243 G substitution is also reported in men with impaired sperm parameters.[67,68]
In an ongoing study in our laboratory, we found increased number of nonsynonymous nucleotide changes in sperm mtDNA of oligoasthenoteratozoospermic (OAT) men as compared with controls;[59] these men had increased ROS levels[62] and also increased nuclear DNA damage.[69] Although much remains to be learnt, a considerable progress has been made in the clinical delineation of genetic forms of male infertility and in the characterization of the responsible genes and their mutations or deletions. It is important to characterize if the genetic etiology is mt or nuclear as cases with mtDNA defects have good prognosis since there is no paternal transmission of mtDNA.
DNA DAMAGE AND INFERTILITY
The sperm chromatin is highly condensed, stable, and is one sixth the size of somatic cell nucleus. It is one transcriptionally inactive. During spermiogenesis, histones are replaced by TP, which subsequently are replaced by PRM,[70,71] which have a high degree of disulfide bonds and make the sperm DNA crystalline. DNA damage manifests as DNA fragmentation and DNA denaturation. Sperm DNA integrity is defined as the absence of DNA nicks or single-/double-stranded breaks and chemical modifications of DNA. The nuclear DNA damage does not impair fertilization or cleavage as paternal genome is activated 72 h postfertilization. Sperm DNA damage results in poor blastocyst development, unequal cleavage, implantation failure, or early fetal loss. Although oocyte repair mechanism repair the damaged sperm DNA, increased DNA damage overwhelms oocyte repair capacity. Certain products of oxidative stress as etheno-nucleosides inhibit oocyte repair capacity by impairing nucleotide excision repair mechanism of oocyte.
Higher degree of negative association between the degree of DNA damage with various indices of fertility as fertilization rate, embryo cleavage rate, implantation rate, pregnancy, and live birth rate have been observed.[72] The spontaneous abortion rate is 1.7-fold higher when more than 30% of sperms contain fragmented DNA. The chances of live birth and conception decreases drastically when DNA fragmentation index is >30.[73]
Infertile men may have sperm with normal sperm parameters, but the germ cells may harbor DNA damage, which cannot be predicted by routine semen analysis. Using these sperm with damaged DNA for ART may lead to increased genetic and epigenetic defects in offspring, childhood malignancies, and increased risk of major and minor congenital malformation.[74] Sperm DNA damage is also one of the major underlying causes of recurrent spontaneous abortions.[75]
Several factors lead to DNA damage, such as oxidative stress,[69,75,76,77] defective chromatin packing, ultracentrifugation, increased histone to protamine ratio, exposure to pollutants, xenobiotics, high temperature and cell phone radiation. The threshold clinical values for the DNA damage beyond which conception is not possible is not optimized yet due to the logistic constrains of the numerous techniques considered. The values are just an approximation depending on the technique adopted. TUNEL, comet, sperm chromatin dispersion, and sperm chromatin structure assay are few of the common techniques. In a study done in our laboratory, we found that in infertile men 40% of the sperms had a very high degree of DNA damage as assessed by increased tail length comet assay.[69,76,77]
Compared with conventional semen analysis, the techniques for DNA damage assessment provide more clinically relevant information with regard to fertility potential of an individual, and these techniques have a very significant role to play in the era of ART where the process of natural selection is bypassed.
mRNA AND INFERTILITY
RNA profiling of fertile and infertile men has shown dynamic cellular variation, and thus has proved to be a biomarker for male infertility. Round spermatids contain numerous varieties of transcripts that are produced either throughout early spermatogenesis or during spermiogenesis from the haploid genome coding for sperm-specific proteins. The transcripts are stored in spermatid cytoplasm before the related proteins are expressed.[78] In midspermiogenesis, the chromatin remodeling leads to the transcriptional inactivation of the genome.[79] Thus the highly condensed sperm nucleus is transcriptionally inert and contains diverse RNA population, mRNA, antisense, and miRNAs that have been transcribed prior to inactivation.[80–83] The presence of transcripts in human spermatozoa has been established using reverse transcription polymerase chain reaction (PCR) and real-time PCR.[84]
The functional significance of spermatozoa RNAs in male fertility was determined by the identification of their role in spermatogenesis, fertilization, and early embryo development. Ostermeier et al. reported that some of these RNAs are transmitted to the oocyte during fertilization.[85] In their later studies, they provided evidence for the presence of interfering RNAs (iRNAs) in spermatozoa.[86] Emery and Carrel,[87] in 2006, reported that altered mRNA profiles contribute to idiopathic male infertility and may affect in vitro fertilization outcome. Six candidate transcripts present only in human spermatozoa and not in oocytes have been identified, which indicates the transfer of RNAs from sperm to oocyte. These RNAs are clusterin, AKAP4 (A-kinase anchoring protein 4), protamine-2, HSBP1 (Heat shock-binding protein 1), FOXG1B (forkhead box G1B), and WNT5A (wingless-type MMTV integration site family).
Clusterin is involved in sperm maturation, lipid transportation, stabilization of stress proteins, and it also influences the oocyte-penetration capacity of cauda epididymal sperm.[88,89] The PRM-2 transcript is degraded in the 2-cell embryo stage.[90] AKAP4 regulates the motility and also plays an important role in cell signaling process.[91] FOXG1B and WNT5A are critical for early embryo patterning,[92] whereas WNT5A is also important for cellular differentiation.[93]
Sperm mRNAs coding for protamines are a useful biomarker for predicting male infertility. Sperm haploinsufficiency for the prm1 and prm2 results in aberrant sperm chromatin condensation and the aberrant prm1/prm2 ratio play an important role in male infertility.[94]
Small RNAs mediate transcriptional gene silencing by methylation[95,96] ; also the miRNA's class of transcripts help in the establishment of imprints in early embryos.[85]
RNA profiles of mature, ejaculated spermatozoa reflect spermatogenic gene expression.[97,98] Gene expression studies by microarray on spermatozoal RNA have shown that mRNA profile of infertile men is distinct from the fertile controls.[99] In a study done by Platts[100] et al., 2007, reported that teratozoospermic men have a distinctive mRNA profile, which can be used in a predictive manner. Thus the differences in the spermatozoa transcriptomes between groups has potential diagnostic and therapeutic possibilities and could be helpful in determining the underlying etiology of male infertility.
CONCLUSION
Male fertility is regulated by a number of genes, which regulate spermatogenesis and are required for DNA recombination, repair, and replication. The maintenance of genomic integrity is of vital importance for germ cells.
Using the advanced approaches of genomics and proteomics and applying the advanced gene expression techniques as microarray would help to interpret the causative mechanisms for infertility. This would not only minimize the risks associated with ART but will also provide an insight into the unexplored aspects of infertility.
Thus not only understanding the role of fertility-related genes is needed but also a deeper insight into the functions of promoters, regulators, and micro-RNA is required.
Although much has been explored to understand the interactive mechanisms associated with fertility, still there are lacunae in our knowledge that need to be filled, so that the treatment of infertility becomes better and a nonempirical approach can be adopted in idiopathic cases. It would also help the ART clinicians, in aiding their patients to produce healthy fertile offspring.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Yoshida A, Miura K, Shirai M. Cytogenetic survey of 1,007 males. Urol Int. 1997;58:166–76. doi: 10.1159/000282975. [DOI] [PubMed] [Google Scholar]
- 2.Dada R, Kumar R, Shamsi MB, Tanwar M, Pathak D, Venkatesh S, et al. Genetic screening in couples experiencing recurrent assisted procreation failure. Ind J Biochem Biophys. 2008;45:116–20. [PubMed] [Google Scholar]
- 3.Dada R, Kumar R, Shamsi MB, Kumar R, Kucheria K, Sharma RK, et al. Higher frequency of Yq microdeletions in sperm DNA as compared to DNA isolated from blood. Asian J Androl. 2007;9:720–2. doi: 10.1111/j.1745-7262.2007.00274.x. [DOI] [PubMed] [Google Scholar]
- 4.Martin RH. Cytogenetic determinants of male fertility. Hum Reprod Update. 2008;14:379–90. doi: 10.1093/humupd/dmn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selice R, Di Mambro A, Garolla A, Ficarra V, Iafrate M, Ferlin A, et al. Spermatogenesis in Klinefelter syndrome. J Endocrinol Invest. 2010;33:789–93. doi: 10.1007/BF03350343. [DOI] [PubMed] [Google Scholar]
- 6.Lim AS, Fong Y, Yu SL. Estimates of sperm sex chromosome disomy and diploidy rates in a 47,XXY/46,XY mosaic Klinefelter patient. Hum Genet. 1999;104:405–9. doi: 10.1007/s004390050975. [DOI] [PubMed] [Google Scholar]
- 7.Kruse R, Guttenbach M, Shartmann B, Schubert R, van der Ven H, Schmid M, et al. Genetic counselling in a patient with XXY/XXXY/XY mosaic Klinefelter's syndrome: estimate of sex chromosome aberrations in sperm before intracytoplasmic sperm injection. Fertil Steril. 1998;69:432–85. doi: 10.1016/s0015-0282(97)00539-6. [DOI] [PubMed] [Google Scholar]
- 8.Rives N, Joly G, Machy A, Siméon N, Leclerc P, Macé B. Assessment of sex chromosome aneuploidy in sperm nuclei from 47,XXY and 46,XY/47,XXY males: comparison with fertile and infertile males with normal karyotype. Mol Hum Reprod. 2000;6:107–12. doi: 10.1093/molehr/6.2.107. [DOI] [PubMed] [Google Scholar]
- 9.Estop AM, Cieply KM, Wakim A, Feingold E. Meiotic products of two reciprocal translocations studied by multicolor fluorescence in situ hybridization. Cytogenet Cell Genet. 1998;83:193–8. doi: 10.1159/000015177. [DOI] [PubMed] [Google Scholar]
- 10.Dada R, Ahmed ME, Talwar R, Kucheria K. Clinical and Genetic study in a XX (SRY negative) male. Int J Med. 2002 [Google Scholar]
- 11.Hamerton JL, Canning N, Ray M, Smith S. A cytogenetic survey of 14,069 newborn infants: incidence of chromosomal abnormalities. Clin Genet. 1975;8:223–43. doi: 10.1111/j.1399-0004.1975.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 12.Kumar R, Shamsi MB, Gaznavi MI, Jena M, Kucheria K, Kumar R, et al. Structural chromosomal anomalies and their association with reproductive failure. Obstet Gynecol Today. 2007;12:152–4. [Google Scholar]
- 13.Meza-Espinoza JP, Anguiano LO, Rivera H. Chromosomal abnormalities in couples with reproductive disorders. Gynecol Obstet Invest. 2008;66:237–40. doi: 10.1159/000147170. [DOI] [PubMed] [Google Scholar]
- 14.Boué A, Gallano P. A collaborative study of the segregation of inherited chromosome structural rearrangements in 1356 prenatal diagnoses. Prenat Diagn. 1984;4:45–67. doi: 10.1002/pd.1970040705. [DOI] [PubMed] [Google Scholar]
- 15.Daniel A, Hook EB, Wulf G. Risks of unbalanced progeny at amniocentesis to carriers of chromosome rearrangements: data from United States and Canadian laboratories. Am J Med Genet. 1989;31:14–53. doi: 10.1002/ajmg.1320330105. [DOI] [PubMed] [Google Scholar]
- 16.Chandley AC, Edmond P, Christie S, Gowans L, Fletcher J, Frackiewicz A, et al. Cytogenetics and infertility in man.II Testicular histology and meiosis. results of a five year survey of men attending a subfertility clinic. Ann Hum Genet. 1976;40:165–76. doi: 10.1111/j.1469-1809.1975.tb00126.x. [DOI] [PubMed] [Google Scholar]
- 17.Hendry WF, Polani PE, Pugh RC, Sommerville IF, Wallace DM. 200 infertile males: correlation of chromosome, histological, endocrine and clinical studies. Br J Urol. 1976;47:899–908. doi: 10.1111/j.1464-410x.1975.tb04074.x. [DOI] [PubMed] [Google Scholar]
- 18.Van Opstal D, Los FJ, Ramlakhan S, Van Hemel JO, Van Den Ouweland AM, Brandenburg H, et al. Determination of the parent of origin in nine cases of prenatally detected chromosome aberrations found after intracytoplasmic sperm injection. Hum Reprod. 1997;12:682–6. doi: 10.1093/humrep/12.4.682. [DOI] [PubMed] [Google Scholar]
- 19.Silber S, Escudero T, Lenahan K, Abdelhadi I, Kilani Z, Munné S. Chromosomal abnormalities in embryos derived from testicular sperm extraction. Fertil Steril. 2003;79:30–8. doi: 10.1016/s0015-0282(02)04407-2. [DOI] [PubMed] [Google Scholar]
- 20.Navarro-Costa P, Gonçalves J, Plancha CE. The AZFc region of the Y chromosome: at the crossroads between genetic diversity and male infertility. Hum Reprod Update. 2010;16:525–42. doi: 10.1093/humupd/dmq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 22.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–24. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 23.Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–43. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 24.Dada R, Gupta NP, Kucheria K. Semen cryopreservation in men with AZFc microdeletion. Clin Genet. 2003;64:74–5. doi: 10.1034/j.1399-0004.2003.00104.x. [DOI] [PubMed] [Google Scholar]
- 25.de Llanos M, Ballescà JL, Gázquez C, Margarit E, Oliva R. High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod. 2005;2:216–20. doi: 10.1093/humrep/deh582. [DOI] [PubMed] [Google Scholar]
- 26.Vogt PH. Genomic heterogeneity and instability of the AZF locus on the human Y chromosome. Mol Cell Endocrinol. 2004;224:1–9. doi: 10.1016/j.mce.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Sharma N, Singh M, Acharya N, Singh SK, Thapa BR, Kaur G, et al. Implication of the cystic fibrosis transmembrane conductance regulator gene in infertile family members of Indian CF patients. Biochem Genet. 2008;46:847–56. doi: 10.1007/s10528-008-9199-x. [DOI] [PubMed] [Google Scholar]
- 28.Kuijper EA, Blankenstein MA, Luttikhof LJ, Roek SJ, Overbeek A, Hompes PG, et al. Frequency distribution of polymorphisms in the FSH receptor gene in infertility patients of different ethnicity. Reprod Biomed Online. 2010;20:588–93. doi: 10.1016/j.rbmo.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Selhub J. Homocysteine metabolism. Annu Rev Nutr. 1999;19:217–46. doi: 10.1146/annurev.nutr.19.1.217. [DOI] [PubMed] [Google Scholar]
- 30.Robaire B, Viger RS. Regulation of epididymal epithelial cell functions. Biol Reprod. 1995;52:226–36. doi: 10.1095/biolreprod52.2.226. [DOI] [PubMed] [Google Scholar]
- 31.Peters M, Saare M, Kaart T, Haller-Kikkatalo K, Lend AK, Punab M, et al. Analysis of Polymorphisms in the SRD5A2 Gene and Semen Parameters in Estonian Men. J Androl. 2009;31:372–8. doi: 10.2164/jandrol.109.008714. [DOI] [PubMed] [Google Scholar]
- 32.Eicheler W, Tuohimaa P, Vilja P, Adermann K, Forssmann WG, Aumüller G. Immunocytochemical localization of human 5 alpha-reductase 2 with polyclonal antibodies in androgen target and non-target human tissues. J Histochem Cytochem. 1994;42:667–75. doi: 10.1177/42.5.8157936. [DOI] [PubMed] [Google Scholar]
- 33.Fernández-Cancio M, Nistal M, Gracia R, Molina MA, Tovar JA, Esteban C, et al. Compound heterozygous mutations in the SRD5A2 gene exon 4 in a male pseudohermaphrodite patient of Chinese origin. J Androl. 2004;25:412–6. doi: 10.1002/j.1939-4640.2004.tb02808.x. [DOI] [PubMed] [Google Scholar]
- 34.Makridakis NM, di Salle E, Reichardt JK. Biochemical and pharmacogenetic dissection of human steroid 5 alpha-reductase type II. Pharmacogenetics. 2000;10:407–13. doi: 10.1097/00008571-200007000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Kojima Y, Mizuno K, Kohri K, Hayashi Y. Advances in molecular genetics of cryptorchidism. Urology. 2009;74:571–8. doi: 10.1016/j.urology.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 36.de Yebra L, Ballescà JL, Vanrell JA, Bassas L, Oliva R. Complete selective absence of protamine P2 in humans. J Biol Chem. 1993;268:10553–7. [PubMed] [Google Scholar]
- 37.Balhorn R, Reed S, Tanphaichitr N. Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia. 1988;44:52–5. doi: 10.1007/BF01960243. [DOI] [PubMed] [Google Scholar]
- 38.Audi L, Fernández-Cancio M, Carrascosa A, Andaluz P, Torán N, Piró C, et al. Novel (60%) and recurrent (40%) androgen receptor gene mutations in a series of 59 patients with a 46,XY disorder of sex development. J Clin Endocrinol Metab. 2010;95:1876–88. doi: 10.1210/jc.2009-2146. [DOI] [PubMed] [Google Scholar]
- 39.Ferlin A, Vinanzi C, Garolla A, Selice R, Zuccarello D, Cazzadore C, et al. Male infertility and androgen receptor gene mutations: clinical features and identification of seven novel mutations. Clin Endocrinol (Oxf) 2006;65:606–10. doi: 10.1111/j.1365-2265.2006.02635.x. [DOI] [PubMed] [Google Scholar]
- 40.Lazaros L, Xita N, Kaponis A, Zikopoulos K, Sofikitis N, Georgiou I. Evidence for sex hormone-binding globulin and androgen receptor genes with semen quality. Andrologia. 2008;40:186–91. doi: 10.1111/j.1439-0272.2008.00835.x. [DOI] [PubMed] [Google Scholar]
- 41.Tut TG, Ghadessy FJ, Trifiro MA, Pinsky L, Yong EL. Long polyglutamine tracts in the androgen receptor are associated with reduced trans-activation, impaired sperm production, and male infertility. J Clin Endocrinol Metab. 1997;82:3777–82. doi: 10.1210/jcem.82.11.4385. [DOI] [PubMed] [Google Scholar]
- 42.Rajpert-De Meyts E, Leffers H, Petersen JH, Andersen AG, Carlsen E, Jørgensen N, et al. CAG repeat length in androgen receptor gene and reproductive variables in fertile and infertile men. Lancet. 2002;359:44–6. doi: 10.1016/s0140-6736(02)07280-x. [DOI] [PubMed] [Google Scholar]
- 43.Dowsing AT, Yong EL, Clark M, McLachlan RI, de Kretser DM, Trounson AO. Linkage between male infertility and trinucleotide repeat expansion in the androgen-receptor gene. Lancet. 1999;354:640–3. doi: 10.1016/s0140-6736(98)08413-x. [DOI] [PubMed] [Google Scholar]
- 44.Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, et al. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci. 2003;116:1847–58. doi: 10.1242/jcs.00391. [DOI] [PubMed] [Google Scholar]
- 45.Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, et al. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol. 2007;27:2582–9. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Malekpour M, Al-Madani N, Kahrizi K, Zanganeh M, Lohr NJ, et al. Sensorineural deafness and male infertility: a contiguous gene deletion syndrome. J Med Genet. 2007;44:233–40. doi: 10.1136/jmg.2006.045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasturi SS, Tannir J, Brannigan RE. The metabolic syndrome and male infertility. J Androl. 2008;29:251–9. doi: 10.2164/jandrol.107.003731. [DOI] [PubMed] [Google Scholar]
- 48.Balhorn R, Cosman M, Thornton K, Krishan VV, Corzett M, Bench G, et al. Protamine mediated condensation of DNA in mammalian sperm. In: Gagnon C, editor. The male gamete: From basic science to clinical applications. Vienna: IL: Cache River Press; 1999. pp. 55–70. [Google Scholar]
- 49.Carrell DT, Liu L. Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities in spermatogenesis. J Androl. 2001;22:604–10. [PubMed] [Google Scholar]
- 50.Corzett M, Mazrimas J, Balhorn R. Protamine 1: Protamine 2 stoichiometry in the sperm of eutherian mammals. Mol Reprod Dev. 2002;61:519–27. doi: 10.1002/mrd.10105. [DOI] [PubMed] [Google Scholar]
- 51.Chevaillier P, Mauro N, Feneux D, Jouannet P, David G. Anomalous protein complement of sperm nuclei in some infertile men. Lancet. 1987;2:806–7. doi: 10.1016/s0140-6736(87)92547-5. [DOI] [PubMed] [Google Scholar]
- 52.Belokopytova IA, Kostyleva EI, Tomilin AN, Vorob’ev VI. Humen male infertility may be due to a decrease of the protamine p2 content in sperm chromatin. Mol Reprod Dev. 1993;34:53–7. doi: 10.1002/mrd.1080340109. [DOI] [PubMed] [Google Scholar]
- 53.Aoki VW, Liu L, Carrell DT. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod. 2005;20:1298–306. doi: 10.1093/humrep/deh798. [DOI] [PubMed] [Google Scholar]
- 54.Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet Genome Res. 2003;103:217–24. doi: 10.1159/000076807. [DOI] [PubMed] [Google Scholar]
- 55.Rousseaux S, Caron C, Govin J, Lestrat C, Faure AK, Khochbin S. Establishment of male-specific epigenetic information. Gene. 2005;345:139–53. doi: 10.1016/j.gene.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 56.van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, van der Vlag J, et al. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol. 2008;8:34. doi: 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klenova EM, Morse HC, 3rd, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 58.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci U S A. 2002;99:6806–11. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar R, Venkatesh S, Kumar M, Tanwar M, Shasmsi MB, Kumar R, et al. Oxidative stress and sperm mitochondrial DNA mutation in idiopathic oligoasthenozoospermic men. Indian J Biochem Biophys. 2009;46:172–7. [PubMed] [Google Scholar]
- 60.Shamsi MB, Kumar R, Bhatt A, Bamezai RN, Kumar R, Gupta NP, et al. Mitochondrial DNA mutations in etiopathogenesis of male infertility. Indian J Urol. 2008;24:150–4. doi: 10.4103/0970-1591.40606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venkatesh S, Deecaraman M, Kumar R, Shamsi MB, Dada R. Role of reactive oxygen species in the pathogenesis of mitochondrial DNA (mtDNA) mutations in male infertility. Indian J Med Res. 2009;129:127–37. [PubMed] [Google Scholar]
- 62.Venkatesh S, Riyaz AM, Shamsi MB, Kumar R, Gupta NP, Mittal S, et al. Clinical significance of reactive oxygen species in semen of infertile Indian men. Andrologia. 2009;41:251–6. doi: 10.1111/j.1439-0272.2009.00943.x. [DOI] [PubMed] [Google Scholar]
- 63.Venkatesh S, Shamsi MB, Kumar R, Gupta NP, Sharma RK, Talwar P, Dada R. Oxidative stress: role of seminal plasma. Obstet Gynecol Today. 2009;14:416–9. [Google Scholar]
- 64.Kao SH, Chao HT, Wei YH. Multiple deletions of mitochondrial DNA are associated with the decline of motility and fertility of human spermatozoa. Mol Hum Reprod. 1998;4:657–66. doi: 10.1093/molehr/4.7.657. [DOI] [PubMed] [Google Scholar]
- 65.Kao S, Chao HT, Wei YH. Mitochondrial deoxyribonucleic acid 4977-bp deletion is associated with diminished fertility and motility of human sperm. Biol Reprod. 1995;52:729–36. doi: 10.1095/biolreprod52.4.729. [DOI] [PubMed] [Google Scholar]
- 66.St John JC, Jokhi RP, Barratt CL. Men with oligoasthenoteratozoospermia harbor higher number of multiple mitochondrial deletions in their spermatozoa, but individual deletions are not indicative of overall etiology. Mol Hum Reprod. 2001;7:103–11. doi: 10.1093/molehr/7.1.103. [DOI] [PubMed] [Google Scholar]
- 67.Thangaraj K, Joshi MB, Reddy AG, Rasalkar AA, Singh L. Sperm mitochondrial mutations as a cause of low sperm motility. J Androl. 2003;24:388–92. doi: 10.1002/j.1939-4640.2003.tb02687.x. [DOI] [PubMed] [Google Scholar]
- 68.Spiropoulos J, Turnbull DM, Chinnery PF. Can mitochondrial mutations cause sperm dysfunction? Mol Hum Reprod. 2002;8:719–21. doi: 10.1093/molehr/8.8.719. [DOI] [PubMed] [Google Scholar]
- 69.Shamsi MB, Venkatesh S, Tanwar M, Talwar P, Sharma RK, Dhawan A, et al. DNA integrity and semen quality in men with low seminal antioxidant level. Mutat Res. 2009;665:29–36. doi: 10.1016/j.mrfmmm.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 70.Rooney AP, Zhang J, Nei M. An unusual form of purifying selection in sperm protein. Mol Biol Evol. 2000;17:278–83. doi: 10.1093/oxfordjournals.molbev.a026307. [DOI] [PubMed] [Google Scholar]
- 71.Filatov MV, Semenova EV, Vorob’eva OA, Leont’eva OA, Drobchenko EA. Relationship between abnormal sperm chromatin packing and IVF results. Mol Hum Reprod. 1999;5:825–30. doi: 10.1093/molehr/5.9.825. [DOI] [PubMed] [Google Scholar]
- 72.Agarwal A, Said TM. Role of sperm chromatin abnormalities and DNA damage in male infertility. Hum Reprod Update. 2003;9:331–45. doi: 10.1093/humupd/dmg027. [DOI] [PubMed] [Google Scholar]
- 73.Evenson DP, Jost LK, Corzett M, Ballhorn R. Sperm chromatin structure assay: its clinical use for detecting sperm DNA fragmentation in male infertility and comparisons with other techniques. J Androl. 2002;23:25–43. doi: 10.1002/j.1939-4640.2002.tb02599.x. [DOI] [PubMed] [Google Scholar]
- 74.Agarwal A, Allamaneni SSR. The effect of sperm DNA damage in assisted reproduction outcomes. Minerva Ginecol. 2004;56:235–45. [PubMed] [Google Scholar]
- 75.Shamsi MB, Venkatesh S, Dhawan A, Singh G, Mukherjee S, Malhotra N, et al. Comet assay: A prognostic tool for DNA integrity assessment in infertile men opting for assisted reproduction. Indian J Med Res. 2010;131:675–681. [PubMed] [Google Scholar]
- 76.Shamsi MB, Venkatesh S, Tanwar M, Talwar P, Sharma RK, Mukherjee S, et al. Sperm DNA integrity analysis and correlation with seminal antioxidant levels in men with idiopathic infertility: Importance of prescreening in couples opting for ART. Obstet Gynecol Today. 2008;12:388–90. [Google Scholar]
- 77.Shamsi MB, Kumar R, Dada R. Evaluation of nuclear DNA damage in human spermatozoa in men opting for assisted reproduction. Indian J Med Res. 2008;127:115–23. [PubMed] [Google Scholar]
- 78.Dadoune JP, Siffroi JP, Alfonsi MF. Transcription in haploid male germ cells. Int Rev Cytol. 2004;237:1–56. doi: 10.1016/S0074-7696(04)37001-4. [DOI] [PubMed] [Google Scholar]
- 79.Kierszenbaum AL, Tres LL. Structural and transcriptional features of the mouse spermatid genome. J Cell Biol. 1975;l65:258–70. doi: 10.1083/jcb.65.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grunewald S, Paasch U, Glander HJ, Anderegg U. Mature human spermatozoa do not transcribe novel RNA. Andrologia. 2005;37:69–71. doi: 10.1111/j.1439-0272.2005.00656.x. [DOI] [PubMed] [Google Scholar]
- 81.Amanai M, Brahmajosyula M, Perry AC. A restricted role for sperm borne micro RNAs in mammalian fertilization. Biol Reprod. 2006;75:877–84. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- 82.Yan W, Morozumi K, Zhang J, Ro S, Park C, Yanagimachi R. Birth of mice after intracytoplasmic injection of single purified sperm nuclei and detection of messenger RNAs and Micro RNAs in the sperm nuclei. Biol Reprod. 2008;78:896–902. doi: 10.1095/biolreprod.107.067033. [DOI] [PubMed] [Google Scholar]
- 83.Krawetz SA. Paternal contribution: New insights and future challenges. Nat Rev Genet. 2005;6:633–42. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 84.Chiang MH, Steuerwald N, Lambert H, Main EK, Steinleitner A. Detection of human leukocyte antigen class I messenger ribonucleic acid transcripts in human spermatozoa via reversetranscription polymerase chain reaction. Fertil Steril. 1994;61:276–80. [PubMed] [Google Scholar]
- 85.Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 86.Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. J Androl. 2005;l26:70–4. [PubMed] [Google Scholar]
- 87.Emery BR, Carrell DT. The effect of epigenetic sperm abnormalities on early embryogenesis. Asian J Androl. 2006;8:131–42. doi: 10.1111/j.1745-7262.2006.00127.x. [DOI] [PubMed] [Google Scholar]
- 88.Trougakos IP, Gonos ES. Clusterin/apolipoprotein J in human aging and cancer. Int J Biochem Cell Biol. 2002;l34:1430–48. doi: 10.1016/s1357-2725(02)00041-9. [DOI] [PubMed] [Google Scholar]
- 89.Moura AA, Chapman DA, Killian GJ. Proteins of the accessory sex glands associated with the oocyte-penetrating capacity of cauda epididymal sperm from Holstein bulls of documented fertility. Mol Reprod Dev. 2007;74:214–22. doi: 10.1002/mrd.20590. [DOI] [PubMed] [Google Scholar]
- 90.Ziyyat A, Lefèvre A. Differential gene expression in pre-implantation embryos from mouse oocytes injected with round spermatids or spermatozoa. Hum Reprod. 2001;16:1449–56. doi: 10.1093/humrep/16.7.1449. [DOI] [PubMed] [Google Scholar]
- 91.Appert-Collin A, Baisamy L, Diviani D. Regulation of g protein- coupled receptor signaling by a kinase anchoring proteins. J Recept Signal Transduct Res. 2006;26:631–46. doi: 10.1080/10799890600923211. [DOI] [PubMed] [Google Scholar]
- 92.Murphy DB, Wiese S, Burfeind P, Schmundt D, Mattei MG, Schulz-Schaeffer W, et al. Human brain factor 1, a new member of the fork head gene family. Genomics. 1994;21:551–7. doi: 10.1006/geno.1994.1313. [DOI] [PubMed] [Google Scholar]
- 93.Moon RT, Brown JD, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–62. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 94.Steger K, Fink L, Failing K, Bohle RM, Kliesch S, Weidner W, et al. Decreased protamine-1 transcript levels in testes from infertile men. Mol Hum Reprod. 2003;9:331–6. doi: 10.1093/molehr/gag041. [DOI] [PubMed] [Google Scholar]
- 95.Castanotto D, Tommasi S, Li M, Li H, Yanow S, Pfeifer GP, et al. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1 A gene promoter in He La cells. Mol Ther. 2005;12:179–83. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 96.Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–62. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miller D. Analysis and significance of messenger RNA in human ejaculated spermatozoa. Mol Reprod Dev. 2000;56:259–64. doi: 10.1002/(SICI)1098-2795(200006)56:2+<259::AID-MRD10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 98.Wang H, Zhou Z, Xu M, Li J, Xiao J, Xu ZY, Sha J. A spermatogenesis related gene expression profile in human spermatozoa and its potential clinical applications. J Mol Med. 2004;82:317–24. doi: 10.1007/s00109-004-0526-3. [DOI] [PubMed] [Google Scholar]
- 99.Garrido N, Martínez-Conejero JA, Jauregui J, Horcajadas JA, Simón C, Remohí J, et al. Micro array analysis in sperm from fertile and infertile men without basic sperm analysis abnormalities reveals a significantly different transcriptome. Fertil Steril. 2009;91:1307–10. doi: 10.1016/j.fertnstert.2008.01.078. [DOI] [PubMed] [Google Scholar]
- 100.Platts AE, Dix DJ, Chemes HE, Thompson KE, Goodrich R, Rockett JC, et al. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum Mol Genet. 2007;16:763–73. doi: 10.1093/hmg/ddm012. [DOI] [PubMed] [Google Scholar]


