Abstract
Cholesterol metabolism is tightly regulated at the cellular level. Here we show that miR-33, an intronic microRNA (miRNA) located within the gene encoding sterol-regulatory element–binding factor–2 (SREBF-2), a transcriptional regulator of cholesterol synthesis, modulates the expression of genes involved in cellular cholesterol transport. In mouse and human cells, miR-33 inhibits the expression of the adenosine triphosphate–binding cassette (ABC) transporter, ABCA1, thereby attenuating cholesterol efflux to apolipoprotein A1. In mouse macrophages, miR-33 also targets ABCG1, reducing cholesterol efflux to nascent high-density lipoprotein (HDL). Lentiviral delivery of miR-33 to mice represses ABCA1 expression in the liver, reducing circulating HDL levels. Conversely, silencing of miR-33 in vivo increases hepatic expression of ABCA1 and plasma HDL levels. Thus, miR-33 appears to regulate both HDL biogenesis in the liver and cellular cholesterol efflux.
Cholesterol is an essential cell membrane component and precursor in metabolic pathways, including steroid hormone and bile acid synthesis. Control of cholesterol levels is essential to human health. In mammalian cells, the sterol-response element–binding protein (SREBP) transcription factors regulate the expression of genes involved in cholesterol biosynthesis and cellular uptake (1). In addition, the liver X receptor (LXR) nuclear hormone receptors are important transcriptional regulators of genes involved in the response to cholesterol excess (2), including those encoding the adenosine triphosphate–binding cassette (ABC) transporters ABCA1 and ABCG1, which promote cellular cholesterol efflux (2). The regulation of these pathways is complex and likely to involve posttranscriptional mechanisms as well.
MicroRNAs (miRNAs) are small (22-nt) endogenous double-stranded RNAs that have emerged as posttranscriptional regulators of physiological processes (3). miRNAs bind to complementary target sites in the 3′ untranslated regions (3′UTRs) of mRNAs, causing translational repression and/or mRNA destabilization (3). A single miRNA can have multiple targets, potentially providing simultaneous regulation of the genes involved in a physiological pathway. We explored whether miRNAs contribute to the maintenance of cholesterol homeostasis.
We undertook an unbiased genome-wide screen of miRNAs modulated by cellular cholesterol content. We identified a subset of 21 miRNAs differentially regulated in human macrophages by cholesterol depletion and cholesterol enrichment, including several whose predicted gene targets are involved in cholesterol uptake, transport, and efflux (table S1). Confirmation of these candidates identified miRNAs that were both up- and down-regulated by cellular cholesterol content (fig. S1A). Sequence alignment revealed that one of these candidates, hsa-miR-33a and its mouse homolog mmu-miR-33 (referred to here as miR-33), is encoded within intron 16 of SREBF2, a gene that encodes a key transcriptional regulator of cholesterol uptake and synthesis (Fig. 1A) (1). Furthermore, the pre-miRNA is highly conserved in mammals (Fig. 1A), prompting us to select miR-33 for further characterization of its role in cholesterol metabolism.
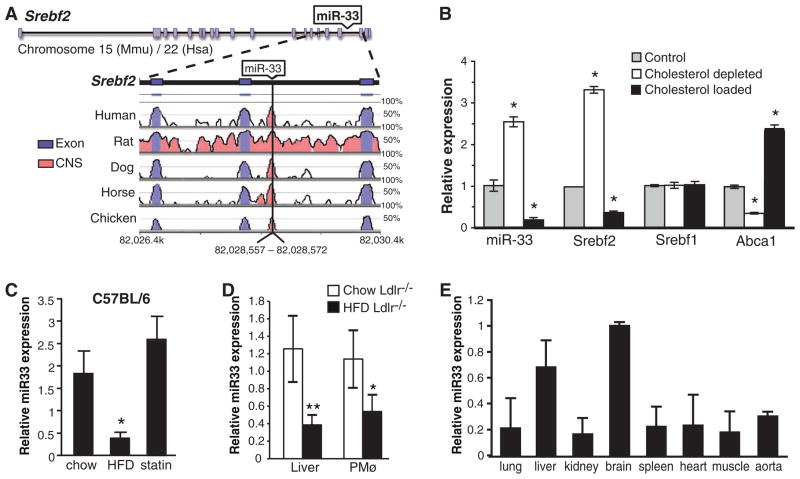
Fig. 1.
Regulation of miR-33 is inversely correlated with cellular cholesterol levels. (A) Schematic representation of the SREBF2 gene locus, demonstrating the miR-33 coding sequence within intron 16 and its conservation among species (reference: the mouse genome). (B) Quantitative real-time fluorescence polymerase chain reaction (QRT-PCR) analysis of miR-33, SREBF2, SREBF1, and ABCA1 expression in control macrophages or macrophages loaded with cholesterol by AcLDL treatment or depleted of cholesterol by statin treatment. *P ≤ 0.05. (C) QRT-PCR analysis of miR-33 in liver of C57BL6 mice (n = 5 per group) fed a chow diet, high-fat diet (HFD), or rosuvastatin-supplemented diet (statin) (D) QRT-PCR analysis of miR-33 in liver and peritoneal macrophages (PMø) from Ldlr−/− mice fed a chow or HFD for 12 weeks (n = 6 per group). **P ≤ 0.01. (E) QRT-PCR analysis of miR-33 tissue expression in C57BL6 mice (n = 3). In (B) to (E), data are the mean ± SEM and are representative of ≥3 experiments.
We found that in mouse peritoneal macrophages, miR-33 and SREBF2 expression were coordinately down-regulated by cholesterol loading, suggesting that these gene regulatory elements are cotranscribed (Fig. 1B). Furthermore, macrophages depleted of cholesterol with the 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitor simvastatin showed robust up-regulation of both miR-33 and SREBF2, but not SREBF1 (Fig. 1B). Levels of miR-33 were inversely correlated with the expression of another cholesterol-responsive gene product, the cholesterol transporter ABCA1 (Fig. 1B). Analysis of the kinetics of miR-33 induction revealed a concomitant increase in miR-33 and SREBP2 levels with simvastatin treatment, consistent with their coregulation (fig. S1B). We next determined whether miR-33 is regulated under physiologic conditions by measuring its expression in mice fed a chow, rosuvastatin-supplemented, or high-fat diet. Consistent with our in vitro observations, hepatic miR-33 levels were inversely correlated with cholesterol levels and ABCA1 expression and positively correlated with SREBF2 mRNA levels, suggesting that miR-33 is regulated by dietary cholesterol in vivo (Fig. 1C and fig. S1, C and D). miR-33 levels were also regulated in two mouse models of hypercholesterolemia: Ldlr−/− and Apoe−/− mice. Hepatic and peritoneal macrophage miR-33 levels were markedly reduced in Ldlr−/− mice that were fed a high-fat diet (Fig. 1D). Similarly, miR-33 levels in peritoneal macrophages from hypercholesterolemic Apoe−/− mice correlated inversely with cellular cholesterol ester content and expression of ABCA1 (fig. S2A). We next examined miR-33 expression in mouse tissues and various cell lines. In addition to macrophages, miR-33 was highly expressed in mouse and human hepatic cells and to a lesser extent in endothelial cells (fig. S2B). Furthermore, miR-33 was widely expressed in mouse tissues and was particularly abundant in the brain and liver (Fig. 1E).
To gain insight into the function of miR-33, we analyzed its potential gene targets, using several miRNA target prediction algorithms (3). We identified putative binding sites for mouse miR-33 in the 3′UTR of several genes involved in cellular cholesterol mobilization, including ABCA1 and ABCG1 and the endolysosomal transport protein NPC1 (fig. S3), suggesting that miR-33 coordinates cholesterol homeostasis through these pathways (4, 5). To test this hypothesis, we determined the effect of miR-33 on the expression of ABCA1, ABCG1, and NPC1 in macrophages treated with acetylated low-density lipoprotein (AcLDL) (to enrich in cholesterol) or the LXR ligand T0901317 (to directly stimulate expression of the three genes) (2). Transfection of mouse peritoneal macrophages with miR-33 (but not a control miRNA) strongly decreased the stimulation of both ABCA1 and ABCG1 protein and mRNA (Fig. 2A and fig. S4A; quantification in fig. S5A). These effects of miR-33 were seen with concentrations of as little as 5 nM (Fig. 2B) and were reversible by co-incubation with an antisense inhibitor of miR-33 (anti-miR-33) (Fig. 2C; quantification in fig. S5, B and C). miR-33 also repressed ABCA1 and ABCG1 protein in mouse hepatic cells, indicating that its effects are not cell type–specific (Fig. 2D). Inhibition of endogenous miR-33 by anti-miR-33 increased the expression of ABCA1 and ABCG1 in macrophages and of ABCA1 in hepatocytes, which is consistent with the hypothesis that miR-33 has a physiological role in regulating the expression of these transporters (Fig. 2E; quantification in fig. S6).
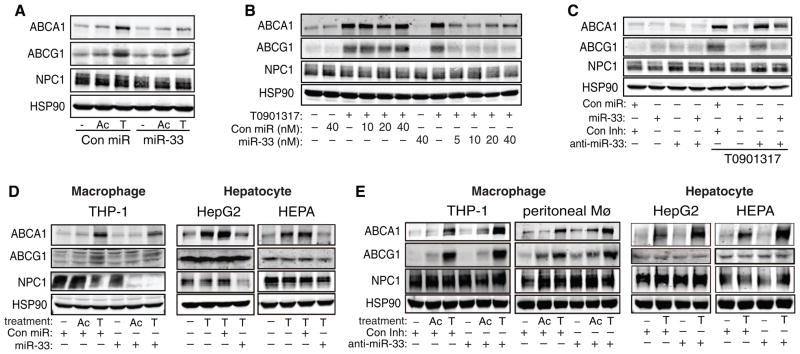
Fig. 2.
Posttranscriptional regulation of ABCA1, ABCG1, and NPC1 by miR-33. (A to C) Western blot analysis of ABCA1, ABCG1, NPC1, and HSP90 in primary mouse macrophages transfected with (A) a control (Con) miR or miR-33, (B) increasing concentrations of control miR or miR-33, or (C) control miR or miR-33 in the presence or absence of a control inhibitor or anti-miR-33. Ac, AcLDL; T, T0901317. (D) Expression of ABCA1, ABCG1, and NPC1 expression in human (THP-1 and HepG2) and mouse (HEPA) cells of the indicated origin transfected with control miR or miR-33. (E) Expression of ABCA1, ABCG1, and NPC1 in human (THP-1 and HepG2) and mouse (peritoneal Mø and HEPA) macrophages and hepatic cells transfected with control inhibitor or anti-miR-33. Data are the mean ± SEM and are representative of ≥3 experiments.
Although we found that miR-33 comparably repressed ABCA1 in cells of mouse and human origin, this was not the case for NPC1 and ABCG1. miR-33 strongly suppressed NPC1 protein in cells of human origin (Fig. 2D), whereas in mouse cells, miR-33 suppressed NPC1 protein levels only modestly and had no effect on NPC1 mRNA levels (fig. S4A). Furthermore, transfection of human macrophage, hepatic, and endothelial cells with miR-33 had no detectable effect on ABCG1 protein (Fig. 2D and fig. S4B). Conservation map analysis revealed that although the predicted miR-33 target sites in the 3′UTR of mouse ABCA1 and NPC1 are highly conserved (fig. S3), the putative sites for miR-33 in the 3′UTR of ABCG1 are present only in mouse and rat. Moreover, a second miR-33–binding site was identified in human NPC1 (fig. S3C), highlighting the species-specific regulation of cholesterol metabolism genes by miR-33.
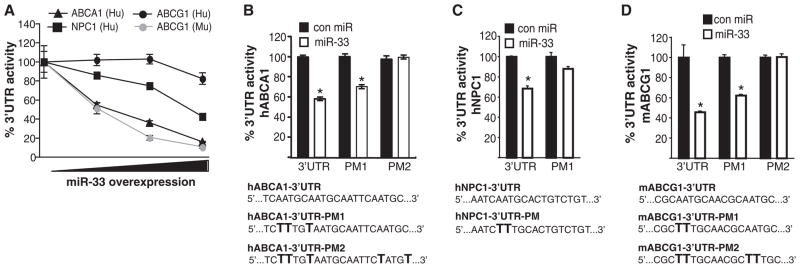
To assess the effects of miR-33 on the 3′UTR of human and mouse ABCA1, ABCG1, and NPC1, we used luciferase reporter constructs. miR-33 markedly repressed mouse, but not human, ABCG1 3′UTR activity (Fig. 3A). Furthermore, consistent with species-conserved miR-33 target sites, miR-33 significantly inhibited human ABCA1 (hABCA1) and hNPC1 3′UTR activity (Fig. 3A). Mutation of the miR-33 target sites relieved miR-33 repression of hABCA1, murine ABCG1 (mABCG1), and hNPC1 3′UTR activity, consistent with a direct interaction of miR-33 with these sites (Fig. 3, B to D). Mutation of both miR-33 sites in the 3′UTR of hABCA1 and mABCG1 was necessary to completely reverse the inhibitory effects of miR-33 (Fig. 3, B and D). miR-33 more strongly repressed hNPC1 as compared to mNPC1 3′UTR activity, which is consistent with the presence of an additional miR-33–binding site (fig. S4C). Together, these experiments identify ABCA1 and NPC1 as conserved targets of miR-33, whereas ABCG1 is a target only in the mouse.
Fig. 3.
miR-33 specifically targets the 3′UTR of ABCA1, NPC1, and mouse, but not human, ABCG1. (A) Activity of luciferase reporter constructs fused to the 3′UTR of human ABCA1, human NPC1, and mouse or human ABCG1 in HEK293 cells transfected with increasing concentrations (0, 5, 50, or 500 ng) of control miR or miR-33. (B to D) Luciferase reporter activity in COS-7 cells transfected with control miR or miR-33 of the (B) hABCA1, (C) hNPC1, and (D) mABCG1 3′UTRs containing the indicated point mutations (PM) in the miR-33 target sites. Data are expressed as mean % of 3′UTR activity of control miR ± SEM and are representative of ≥3 experiments. *P ≤ 0.05.
To confirm the specificity of miR-33 targeting of ABCA1, ABCG1, and NPC1, we assessed the effect of miR-33 overexpression on other lipid metabolism–related genes in macrophages and hepatocytes. Whereas ABCA1 was predictably down-regulated in these cells, we observed few changes in the expression of non–miR-33 targets (fig. S7, A and B). Other genes containing putative miR-33–binding sites such as HMGCR and SCAP were modestly down-regulated at the mRNA level, but there was no detectable change in protein expression (fig. S8).
The ability of ABCA1 and ABCG1 to stimulate the efflux of cholesterol from cells in the periphery, particularly cholesterol-laden macrophages in atherosclerotic plaques, is an important antiatherosclerotic mechanism (5). Transfection of J774 murine macrophages with miR-33 attenuated cholesterol efflux to apolipoprotein A1 (apoA1) and high-density lipoprotein (HDL), in agreement with the known functions of ABCA1 and ABCG1, respectively (Fig. 4, A and B). miR-33 did not impair cholesterol efflux to HDL in human THP-1 macrophages, which is consistent with the lack of miR-33–binding sites in the human ABCG1 3′UTR (Fig. 4B). Similar results were seen in human and rat hepatocytes, where transfection of miR-33 reduced cholesterol efflux to apoA1 (fig. S9A). Antagonism of endogenous miR-33 increased ABCA1 protein and cholesterol efflux to apoA1 in both murine and human macrophages (Fig. 4C and fig. S9B). Under conditions in which miR-33 is increased (cholesterol depletion), anti-miR-33 significantly increased cholesterol efflux in both macrophages and hepatocytes (fig. S9C). Thus, manipulation of cellular miR-33 levels alters macrophage cholesterol efflux, a critical first step in the reverse cholesterol transport pathway for the delivery of excess cholesterol to the liver (5).
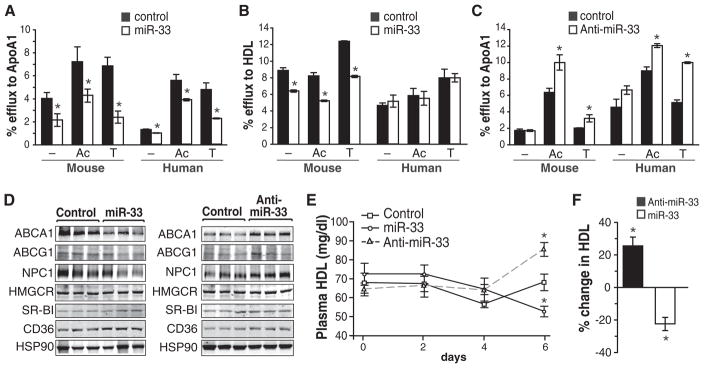
Fig. 4.
Modulation of miR-33 regulates cellular cholesterol efflux and serum HDL levels. (A and B) Cholesterol efflux to (A) apoA1 and (B) HDL in mouse J774 and human THP-1 macrophages stimulated with AcLDL (Ac) or T0901317 (T) and expressing a control miR or miR-33. (C) Cholesterol efflux to apoA1 in mouse J774 and human THP-1 macrophages stimulated with AcLDL or T0901317 and expressing a control inhibitor or anti-miR-33. (D) Analysis of hepatic gene expression 6 days after infection with the control, miR-33, or anti-miR-33 lentiviruses. Western blots are of liver tissue from three representative mice per treatment. (E) Plasma HDL levels in mice infected with the control, miR-33, or anti-miR-33 lentiviruses. (n = 6). (F) Percentage change in serum HDL 6 days after lentiviral delivery of miR-33 or anti-miR-33 (relative to control). Data are the mean ± SEM. *P ≤ 0.05
In addition to cellular cholesterol efflux, ABCA1 is responsible for initiating HDL formation in the liver (6). Thus, we investigated the effect of manipulating miR-33 levels in vivo in mice using lentiviruses encoding pre-miR-33, anti-miR-33, or control. Efficient lentiviral delivery was confirmed by measuring green fluorescent protein in the liver (fig. S9D). Consistent with our in vitro results, miR-33 significantly reduced hepatic ABCA1 expression (Fig. 4D; quantification in fig. S9E). It also modestly decreased ABCG1 and NPC1 protein levels, although the effect was not statistically significant. No changes in SR-B1, a cognate receptor for HDL in the liver (7), or other cholesterol-related genes were observed (Fig. 4D). Moreover, an unbiased assessment of hepatic gene expression revealed few significant differences in the expression of other cholesterol metabolism–related genes in mice treated with miR-33 or anti-miR-33 lentiviruses (fig. S10). In vivo overexpression of miR-33 resulted in a progressive decline of plasma HDL, as expected from the requirement of ABCA1 for HDL formation (Fig. 4E), with a 22% decrease achieved after 6 days (Fig. 4F). Conversely, mice expressing anti-miR-33 showed a 50% increase in hepatic ABCA1 protein and a concomitant 25% increase in plasma HDL levels after 6 days (Fig. 4, D to F). Thus, manipulation of miR-33 levels in vivo alters ABCA1 expression and the mobilization of cholesterol to HDL.
To date, only one other miRNA, miR-122, has been shown to have a direct role in cholesterol metabolism (8–10). The expression of miR-122 is highly restricted to the liver, where it is believed to maintain the differentiated state. Silencing of miR-122 down-regulates genes implicated in cholesterol biosynthesis and tri-glyceride metabolism, increasing hepatic fatty acid oxidation and reducing plasma cholesterol, hepatic fatty acid, and cholesterol synthesis. In contrast, miR-33 is widely expressed in different cell types and tissues, consistent with the hypothesis that it has a more global effect on cellular cholesterol homeostasis. Moreover, we have shown that miR-33 specifically regulates cholesterol transport pathways that mobilize cholesterol from intracellular stores to HDL lipoproteins.
Although the pathways regulating the generation and uptake of LDL cholesterol are well characterized, the molecular mechanisms regulating circulating levels of HDL, the “good cholesterol,” remain poorly defined. The identification of ABCA1 as the gene mutated in Tangier disease, a condition characterized by a near-deficiency of plasma HDL, revealed its essential role in both HDL generation and reverse cholesterol transport (11–13). Subsequent studies have established that ABCA1 and ABCG1 probably act in a sequential fashion, with ABCA1 lipidating apoA1 to generate nascent HDL particles, which can then promote additional cholesterol efflux via ABCG1 (5). Despite these major advances, it has become clear that the regulation of these pathways is complex and influenced not only by genetic factors but also by posttranscriptional mechanisms (14). Our data provide evidence for a role for miR-33 in the epigenetic regulation of cholesterol homeostasis. We propose that miR-33 functions via a negative feedback loop triggered by the cholesterol content of the cell; under low sterol conditions, the coincident transcription of SREBF2 and miR-33 coordinate cellular cholesterol homeostasis by simultaneously initiating transcription of cholesterol uptake and synthesis pathways and posttranscriptionally repressing genes involved in cellular cholesterol export.
Our work identifies miR-33 as a potential regulator of two central pathways that control HDL cholesterol: (i) HDL biogenesis in the liver, as reflected by the impact of miR-33 manipulation on circulating HDL levels; and (ii) cellular cholesterol efflux from macrophages, the first step in the reverse cholesterol transport pathway through which HDL and apoA1 ferry excess cholesterol back to the liver for excretion. Because plasma HDL levels show a strong inverse correlation with atherosclerotic vascular disease, there has been intense interest in therapeutically targeting HDL and macrophage cholesterol efflux pathways. Our study suggests that antagonists of endogenous miR-33 may be a useful therapeutic strategy for enhancing ABCA1 expression and raising HDL levels in vivo.
Supplementary Material
Acknowledgments
We thank E. Hernando-Monje for assisting with lentiviral experiments and M. Freeman and L. Stuart for discussions. This work was supported by the American Heart Association (grant SDG-0835585D to C.F.-H., grant SDG-0835481N to Y.S., and grant 09GRNT2260352 to M.L.F.); NIH (grant R01AG02055 to K.J.M., grant R01HL074136 to M.L.F., grant R01HL084312 to E.A.F., and grant 1P30HL101270-01 to C.F.-H.); and the Heart and Stroke Foundation of Canada (K.J.R.). The authors (C.F.-H. and K.J.M) and New York University School of Medicine are preparing a patent application relating to the use of miR-33 as a therapeutic.
Footnotes
References and Notes
- 1.Horton JD, Goldstein JL, Brown MS. J Clin Invest. 2002;109:1125. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaven SW, Tontonoz P. Annu Rev Med. 2006;57:313. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ory DS. Trends Cardiovasc Med. 2004;14:66. doi: 10.1016/j.tcm.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. Cell Metab. 2008;7:365. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Oram JF, Vaughan AM. Curr Opin Lipidol. 2000;11:253. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Krieger M. J Clin Invest. 2001;108:793. doi: 10.1172/JCI14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elmén J, et al. Nature. 2008;452:896. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 9.Esau C, et al. Cell Metab. 2006;3:87. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Krützfeldt J, et al. Nature. 2005;438:685. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 11.Bodzioch M, et al. Nat Genet. 1999;22:347. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 12.Brooks-Wilson A, et al. Nat Genet. 1999;22:336. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 13.Rust S, et al. Nat Genet. 1999;22:352. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 14.Wellington CL, et al. Lab Invest. 2002;82:273. doi: 10.1038/labinvest.3780421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.