Abstract
The present study screened riboflavin mimicking small molecules to determine their binding activity for the riboflavin binding protein. We performed thermodynamic and kinetic binding studies of these molecules using a combination of two analytical approaches: isothermal titration calorimetry and surface plasmon resonance spectroscopy. Screening of a biased set of nonriboflavin-based small molecules by microcalorimetry led to the discovery of two known drug molecules, quinacrine and chloroquine, as favorable ligands for the riboflavin receptor with KD values of 264 and 2100 nM, respectively. We further demonstrated that quinacrine is a competitive ligand for the receptor as measured by surface plasmon resonance. Thus, this study describes the identification of a novel class of dual-acting riboflavin antagonists that target riboflavin receptor for cellular uptake and display multifunctional activities upon cellular entry.
Keywords: Targeted delivery, riboflavin antagonist, isothermal titration calorimetry, surface plasmon resonance spectroscopy
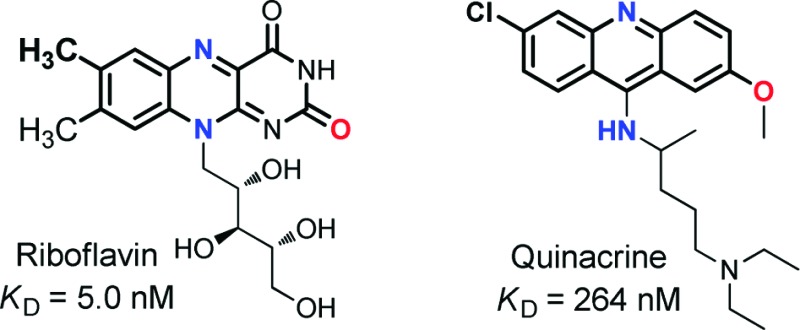
Water-soluble riboflavin (RF, Figure 1) is an essential vitamin (B2) necessary for the biosynthesis of flavin-based redox cofactors, such as flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD). The cellular availability of this vitamin for cofactor biosynthesis is mediated by RF receptors that facilitate the uptake of the RF molecules with high efficiency.1 The RF receptors, also referred to as RF carriers, are expressed as both soluble and membrane-bound isoforms.1 We have recently begun to explore RF as a ligand for targeting cancer therapy for a number of reasons. First, RF receptors are identified to be overexpressed in certain human cell lines from breast and prostate cancers, potentially making this family of proteins a type of tumor biomarker.2,3 Second, recent studies demonstrated that these membrane-bound receptors mediate the cellular uptake of natural RF and RF conjugates by receptor-mediated endocytosis.4−6 On the basis of these studies, we successfully demonstrated receptor-mediated RF-targeted drug delivery to cancer cells in vitro.7 In that study, RF-conjugated dendrimer nanoparticles delivered the anticancer drug methotrexate (MTX) to kill KB cells via receptor-mediated internalization. This work suggested a general route for the selective delivery of anticancer drug molecules to the cancer cells that overexpress RF receptors.
Figure 1.
Structures of RF, RF-linker (1), and heterocycle-based small molecules evaluated for their potential activity as the ligands for RF receptor.
The present study reports the identification and characterization of novel, dual-acting RF antagonists that should make immediate impacts on RF receptor-mediated cancer targeting. By definition, RF antagonists refer to a class of small molecules that are structurally related to RF and interfere with the cellular activity of RF by competitively binding to RF receptors or enzymes whose activities are associated with flavin cofactors.8,9 We sought to identify dual-acting RF antagonists due to their potential benefits for targeted delivery in cancer. These RF antagonists should be able to bind to the RF receptor to mediate uptake via the internalization of the complex formed with a RF receptor. However, unlike native RF that has positive trophic effects on cancer, these molecules can kill cancer cells by interfering with the cellular functions of flavin cofactors such as FMN and FAD, which are essential for cellular maintenance.9,10 Thus, a RF antagonist has the potential to function as a dual-acting ligand in cell targeting.
In an effort to develop RF antagonists for targeted drug delivery applications, we tried to identify new RF antagonists amenable for chemical modifications to allow conjugation to nanoparticles and have the potential to induce cytotoxicity. To identify such potential RF antagonists, we searched in the SciFinder Scholar database focusing on small drug molecules whose core structures mimic the isoalloxazine head of RF. The rationale for this search is that the isoalloxazine heterocycle makes a key contribution for the receptor binding by stacking between the two hydrophobic planes that comprise a receptor cleft for mammalian RF binding protein (RfBP).11 Our structural search led to a small set of compounds comprised of lumiflavin, perphenazine, chlorpromazine, quinacrine, and chloroquine (Figure 1). Of these, quinacrine and chlorpromazine have been previously investigated by circular dichroism (CD) and fluorescence spectroscopy for the receptor binding, but no evidence was found for stereospecific binding of quinacrine and chlorpromazine, while that study did not rule out the possibility for complex formation with RfBP.12 Each of the selected molecules contains a flat heterocycle involving a H-bond acceptor at the middle of the fused heterocycle, such as pyridine (N) or thiazine (S) instead of the pyrazine (N) for RF. This study screens these molecules to determine their binding activity to RfBP using two well-established methods involving isothermal titration calorimetry (ITC) and surface plasmon resonance (SPR). The binding experiments are performed with chicken RfBP, a highly validated RF receptor displaying the significant level of interspecies homology in structure and function,1,8,11 and its results are reported here along with the thermodynamic and kinetic characteristics of the interaction of each of these small molecules with RfBP.
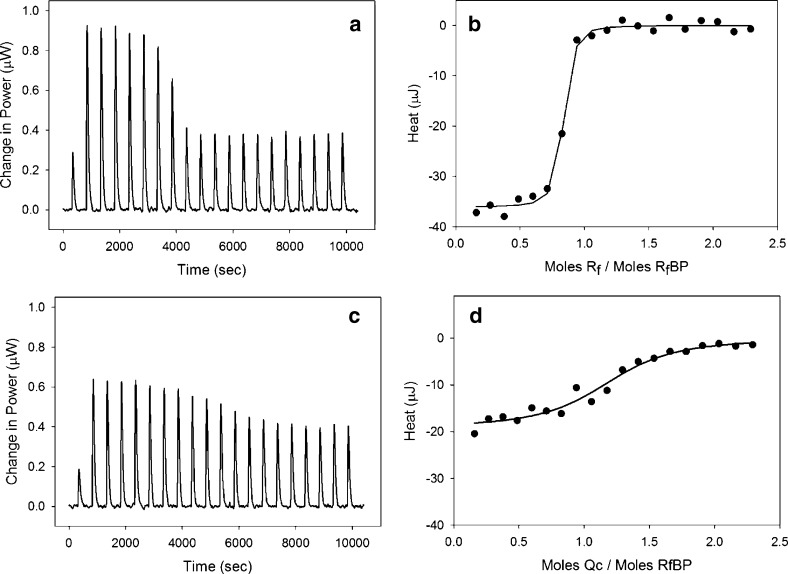
ITC was first used for determining the dissociation constant and the binding stoichiometry for the interaction between RF and RfBP at pH 4, 7.4, and 9. Figure 2 shows the ITC thermograms and the corresponding integrated area of the peaks for RF and quinacrine binding to RfBP in PBS (0.1 M sodium phosphate, 0.1 M NaCl, pH 7.4). The data in Figure 2b,d were fitted to an independent model, and the model fit parameters ΔH, KA, and n (binding stoichiometry) are shown in Table 1, as are the calculated estimates for ΔG and ΔS. The nanomolar binding constants obtained for a pH range spanning five orders of magnitude suggests a tight binding between RF and RfBP. The binding is optimal at physiological pH. However, the affinity is significantly lower at pH 4, an observation supportive of the mechanism of pH-mediated RF dissociation in endosomes (pH ∼ 5).6,8 The large ΔΗ and the smaller ΔS values indicate that the binding interaction is largely enthalpy driven. The apparent dissociation constant, KD of 6.6 nM in 0.1 M phosphate from ITC, is in excellent agreement with two previous studies involving fluorescence quenching, where a KD of 2 nM was obtained in 0.1 M phosphate buffer13 and a KD of 1.3 nM was obtained in 0.01 M phosphate buffer for the RF−RfBP binding interaction.8
Figure 2.
Raw ITC data for the interaction between (a) RF (40 μM) and (c) quinacrine (40 μM) with chicken RfBP (4 μM) at 25 °C in PBS buffer. Plot of integrated area under each injection peak for RF (b) and quinacrine (d). The solid line (b and d) is an independent model fit to data with parameters n, KA, and ΔH.
Table 1. Thermodynamic Parameters Showing pH Dependence of the RF System at 25 °C.
| system | pH | na | KDa (nM) | ΔHa (kJ mol−1) | ΔG (kJ mol−1) | ΔS (kJ mol−1 K−1) |
|---|---|---|---|---|---|---|
| RF vs RfBP (0.1 M phosphate) | 7.4 | 0.81 ± 0.03 | 6.6 ± 1.8 | −82.2 ± 5.7 | −46.6 | −0.12 |
| RF vs RfBP (PBS) | 7.4 | 0.78 ± 0.02 | 5.0 ± 0.7 | −91.2 ± 5.7 | −47.5 | −0.15 |
| RF vs RfBP (0.1 M NaAc) | 4.0 | 0.70 ± 0.04 | 46 ± 2.8 | −100.7 ± 10.4 | −41.8 | −0.20 |
| RF vs RfBP (0.1 M Tris) | 9.0 | 0.82 ± 0.03 | 34 ± 3.0 | −99.1 ± 9.0 | −42.6 | −0.19 |
Reported errors are from fitting data. Error observed between repeats of experiments is similar. KD = 1/KA.
The binding of the five potential RF antagonists to RfBP was examined by ITC. A representative binding isotherm is shown for quinacrine with RfBP in Figure 2c,d, and the thermodynamic parameters from the binding of the RF antagonists are shown in Table 2. On the basis of KD, the order of binding strength for RF and RF antagonists to RfBP is RF > lumiflavin > quinacrine > chloroquine. The order of the binding affinities is determined primarily by the magnitude of the enthalpic changes (exothermic), but it is also influenced by the entropic term especially for those molecules where the enthalpic contribution is similar (e.g., lumiflavin vs quinacrine). Table 2 also shows that ΔH and ΔG values are closer in magnitude for RF analogues, suggesting that they do not fully replicate the intermolecular contacts between RF and RfBP. The differences in binding entropy are likely due to protein conformational changes upon binding to RF, which do not occur for RF analogues. Other tested compounds, perphenazine and chlorpromazine, showed no binding affinity toward the RfBP. Although compounds were selected by structural similarities based on the tricyclic heterocycle, perphenazine and chlorpromazine are the only two molecules where a sulfur atom (S) has replaced the N at the N(5) position in the isoalloxazine ring. The ITC studies indicate that changes to the N(5) position inhibit the binding of RF antagonists to RfBP. Because the N(5) position is involved in redox reactions, it is possible that changes at this position affect binding to RfBP.1,11 In addition, the ability of N to serve as the H-bond acceptor may be critical for the tighter binding, while replacement with S can eliminate such ability. Lumiflavin, quinacrine, and chloroquine show nearly 1−3 orders of magnitude in change in KD. The major structural difference among these three RF antagonists is the substituent at position N(10). Clearly, replacement with a nonribose side chain has an effect of increasing the KD values by 1−2 orders of magnitude in comparison to RF. Furthermore, a reduction in the binding affinity is achieved by replacing the alloxazine ring on the heterocycle head of RF. Thus, by screening several RF-mimicking molecules with polarity and structural similarities to RF, we have demonstrated that binding affinity of RF antagonists toward RfBP is tunable. While a prior study based on CD spectroscopy led to an inconclusive result for quinacrine and chlorpromazine,12 our study shows broad applicability and sensitivity of the ITC method for studies involving RF receptor−ligand interactions.14 This method enables us to clearly demonstrate the binding of quinacrine to RfBP at the submicromolar concentration and to confirm the lack of evidence found for the receptor binding with chlorpromazine.
Table 2. Thermodynamic Parameters of RF and RF Antagonists Binding with RfBP in PBS (pH 7.4) at 25 °C.
| system | na | KDa (nM) | ΔHa (kJ mol−1) | ΔG (kJ mol−1) | ΔS (kJ mol−1 K−1) |
|---|---|---|---|---|---|
| RF vs RfBP | 0.78 ± 0.02 | 5.0 ± 0.7 | −91.2 ± 5.7 | −47.5 | −0.15 |
| lumiflavin vs RfBP | 1.08 ± 0.07 | 61 ± 3.5 | −48.2 ± 7.2 | −41.2 | −0.02 |
| quinacrine vs RfBP | 0.90 ± 0.04 | 264 ± 22.1 | −51.6 ± 3.9 | −37.5 | −0.05 |
| chloroquine vs RfBP | 1.06 ± 0.04 | 2100 ± 180 | −40.4 ± 2.9 | −32.4 | −0.03 |
| perphenazine vs RfBP | no binding observed | ||||
| chlorpromazine vs RfBP | no binding observed | ||||
See Table 1.
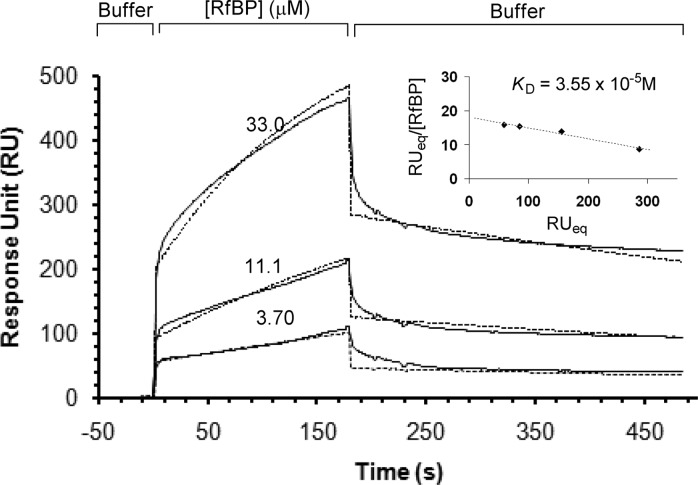
On the basis of the ITC studies, we chose quinacrine with a KD of 0.26 μM for further study by SPR spectroscopy. The present SPR study for RfBP−ligand interactions is based on an approach that utilizes the presentation of an RF ligand to the surface of an SPR sensor chip prepared by immobilization of RF-linker 1 (Scheme S1 in the Supporting Information).15 Binding of RfBP to the RF-presenting sensor chip was studied by varying the concentrations of the protein in the range of 33.0 to 3.70 μM (Figure 3). RfBP bound to the surface in a dose-dependent manner characterized by the biphasic-like association, noticeably at higher doses. Such binding features might reflect various levels of accessibility to the hydrated dextran surface presenting RF ligands or conformational changes of the protein upon ligand occupation. The binding results also suggest slow dissociation of the protein, a feature reported earlier.15 As a control, bovine serum albumin did not bind to the RF-presenting surface in a specific manner (Figure S3 in the Supporting Information). The amount of RfBP bound at the surface was analyzed by the Scatchard plot as a function of the concentrations of RfBP in the buffer (inset in Figure 3). The analysis gave a value for the equilibrium dissociation constant KD (3.55 × 10−5 M). The dissociation constant for RfBP was also calculated by a different method of analysis, where we calculated kinetic binding parameters (off rate = koff; on rate = kon) by analyzing each sensorgram according to BIAevaluation software. Given the 1:1 stoichiometry for the receptor−ligand interaction,8,15 we utilized the Langmuir model16 for the global curve fitting (dotted lines in Figure 3) and derived the kinetic parameters (kon = 1.03 × 102 M−1 s−1; koff = 1.49 × 10−3 s−1; χ2 = 2.5). The resulting equilibrium dissociation constant KD was determined to be 1.45 (±0.49) × 10−5 M. This KD value obtained from the kinetic analysis is close to the value (KD = 3.55 × 10−5 M) determined from the Scatchard analysis for RfBP binding.
Figure 3.
Binding of RfBP to the ligand 1 immobilized to a CM5 sensor chip surface. The concentration of RfBP in the HBS-EP buffer (pH 7.4) is indicated on each of the sensorgrams. The SPR sensorgrams were corrected for bulk effect (Figure S1 in the Supporting Information; global fitting curves are shown in dotted lines). The KD was determined by the Scatchard plot as shown in the inset where RUeq refers to the response unit (RU) when the protein binding reaches equilibrium on the surface.
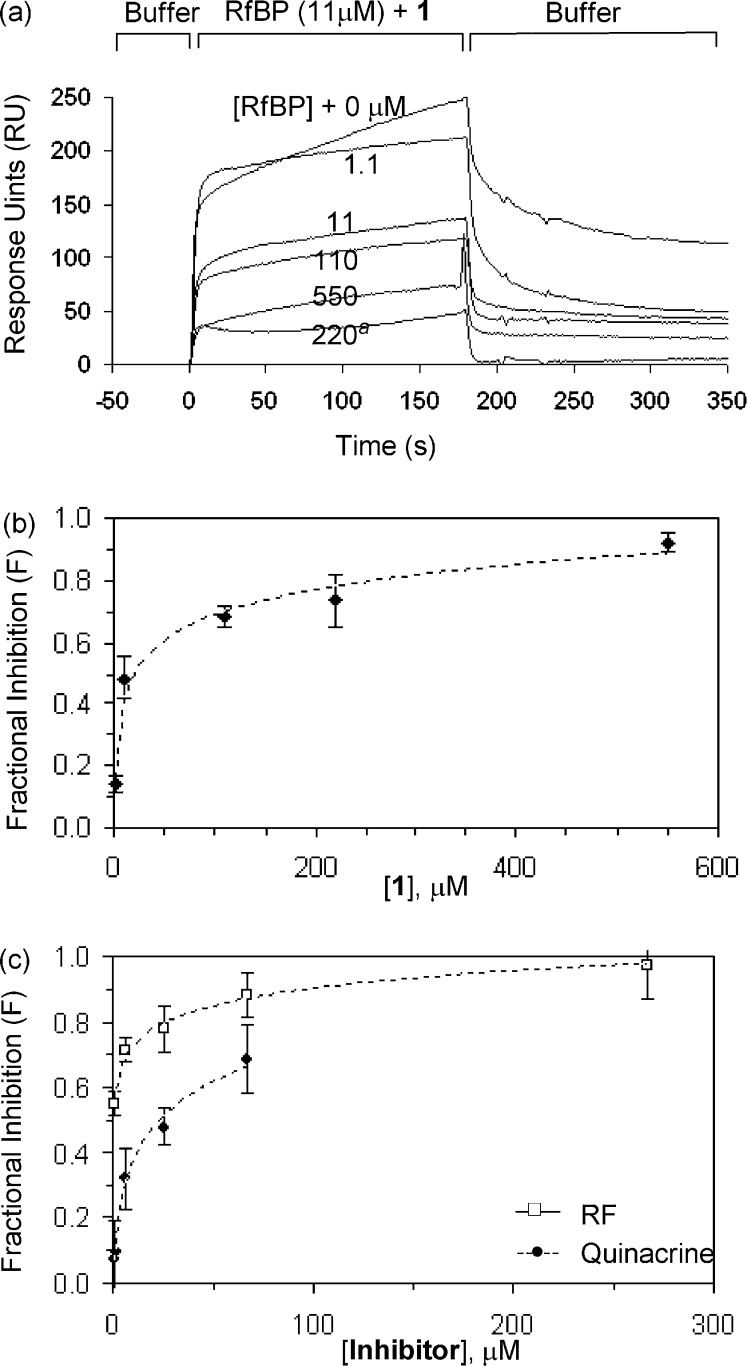
The SPR method developed here was then used to screen RF-mimicking small molecules for inhibiting the adsorption of RfBP to the chip surface presenting RF ligand 1. As an illustration, Figure 4 shows that the addition of soluble RF ligand 1 to the RfBP-containing solution inhibited the binding of RfBP (11.1 μM) to the surface. The amount of RfBP adsorbed to the surface decreased with increasing concentrations of 1 in solution in a dose-dependent manner where approximately 90% of the protein adsorption was inhibited at the concentrations above ∼500 μM. The results from the competition experiments suggest that the interaction between RfBP and 1 immobilized at the surface is biospecific and can be competitively blocked by adding free 1 in the solution. The ligand competition experiment was also performed with RF and quinacrine, the RF-mimicking small molecule identified as the ligand for RfBP from the ITC study (Figure 4 and Figure S2 in the Supporting Information). Each of the experiments suggests for specific blocking of RfBP adsorption to the surface as a function of the ligand concentration. On the basis of the SPR results, the inhibition constant Ki was derived for each ligand according to the solution competition equation,17 and its values are summarized in Table S2 in the Supporting Information. As a reference molecule, RF has a Ki value of 0.35 μM, an inhibitory activity approximately 7-fold more potent than its N(3) derivative 1. In addition, the SPR experiments demonstrated that quinacrine is a competitive inhibitor for RfBP with a Ki value of 6.7 μM. Thus, quinacrine has an inhibitory activity for RfBP ∼19-fold lower than RF.
Figure 4.
(a) Representative SPR sensorgrams from the competitive binding experiments with 1. The binding of RfBP (11.1 μM) to the RF (1)-immobilized chip surface was competitively inhibited by the addition of the soluble ligand 1 to the RfBP solution. The concentrations of the added RF ligand are indicated in the overlay of the sensorgrams. aAn example of the control experiment performed by the injection of 1 alone (220 μM) without RfBP. Each of the RfBP sensorgrams (RfBP + ligand) was corrected against the contribution by the soluble ligand (ligand alone) measured at the designated concentration. (b and c) Plot of fractional inhibition (F = 1 − RU[I]/RU[I]=0) as a function of inhibitor (ligand) concentration for each of the competitive inhibitors (RF, quinacrine, and 1). The response unit (RU[I]) for each sensorgram was determined by correcting the bulk contribution as described earlier. The range of inhibition concentrations was lower in the case of quinacrine due to its limited solubility in the SPR running buffer.
In summary, the SPR results are in good agreement with the findings from the ITC study. It also confirms that quinacrine and chloroquine are newly identified members of RF-mimicking competitive ligands. As drugs traditionally used in the treatment of malaria18 and rheumatoid arthritis,19 these molecules are reported to display diverse biological activities due to the ability to inhibit a number of important biological targets such as DNA topoisomerase II18,20 and metabolic enzymes.21 Recently, quinacrine has generated new attention because of the discovery that it has antitumoral activity.22−25 This activity is attributed to its ability to interfere with cell signaling pathways such as activation of the p53 pathway25 and inhibition of Bcl-xL, an antiapoptotic protein.23 The present study suggests another novel application for the quinacrine class of the drug molecules as ligands that can target RfBP, a vitamin B2 uptake receptor, in a manner competitive to RF.
Implications for this finding are many-fold. First, the present study suggests a new additional perspective for the biological activities associated with quinacrine and its analogues. On the basis of the interpretation of our data, it is also conceivable that quinacrine can interfere with receptor-mediated RF uptake outside the cell and/or can block a broad range of flavin cofactor-mediated enzymatic activities after internalization. Second, despite having a moderate affinity to RF receptor at the lower micromolar concentration, quinacrine can serve as a targeting ligand for specific delivery of additional therapeutic molecules or imaging agents to the receptor-overexpressing cancer cells implicated in breast and prostate cancers.2,3 Pertinent to this targeting utility, it would be possible to apply the concept of multivalent ligand design,7,16,26 in which even suboptimal targeting capability can be enhanced through multivalent tight binding.
K.S. thanks the support of NIH (1F33CA138031-01A1), NSF (CHE-0959681), and HHMI (student support). Part of this work was supported by NCI, NIH under award 1 R01 CA119409 (J.R.B.).
Supporting Information Available
Experimental details and additional SPR sensorgrams. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- White H. B.; Merrill A. H. Riboflavin-Binding Proteins. Annu. Rev. Nutr. 1988, 81279–299. [DOI] [PubMed] [Google Scholar]
- Karande A. A.; Sridhar L.; Gopinath K. S.; Adiga P. R. Riboflavin carrier protein: A serum and tissue marker for breast carcinoma. Int. J. Cancer (Pred. Oncol.) 2001, 95, 277–281. [DOI] [PubMed] [Google Scholar]
- Johnson T.; Ouhtit A.; Gaur R.; Fernando A.; Schwarzenberger P.; Su J.; Ismail M. F.; El-Sayyad H. I.; Karande A.; Elmageed Z. A.; Rao P.; Raj M. Biochemical characterization of riboflavin carrier protein (RCP) in prostate cancer. Front. Biosci. 2009, 14, 3534–3640. [DOI] [PubMed] [Google Scholar]
- Phelps M. A.; Foraker A. B.; Gao W.; Dalton J. T.; Swaan P. W. A Novel Rhodamine-Riboflavin Conjugate Probe Exhibits Distinct Fluorescence Resonance Energy Transfer that Enables Riboflavin Trafficking and Subcellular Localization Studies. Mol. Pharm. 2004, 1, 257–266. [DOI] [PubMed] [Google Scholar]
- Holladay S. R.; Yang Z.-f.; Kennedy M. D.; Leamon C. P.; Lee R. J.; Jayamani M.; Mason T.; Low P. S. Riboflavin-mediated delivery of a macromolecule into cultured human cells. Biochim. Biophys. Acta, Gen. Subj. 1999, 14261195–204. [DOI] [PubMed] [Google Scholar]
- Huang S.-N.; Phelps M. A.; Swaan P. W. Involvement of Endocytic Organelles in the Subcellular Trafficking and Localization of Riboflavin. J. Pharmacol. Exp. Ther. 2003, 3062681–687. [DOI] [PubMed] [Google Scholar]
- Thomas T. P.; Choi S. K.; Li M.-H.; Kotlyar A.; Baker J. R. Design of Riboflavin-presenting PAMAM Dendrimers as a New Nanoplatform for Cancer-targeted Delivery. Bioorg. Med. Chem. Lett. 2010, 20, 5191–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becvar J.; Palmer G. The binding of flavin derivatives to the riboflavin-binding protein of egg white. A kinetic and thermodynamic study. J. Biol. Chem. 1982, 257105607–5617. [PubMed] [Google Scholar]
- Chu C. K.; Bardos T. J. Synthesis and inhibition analysis of 2(4)-imino-4(2)-amino-2,4-dideoxyriboflavin, a dual antagonist of riboflavin and folinic acid. J. Med. Chem. 1977, 202312–314. [DOI] [PubMed] [Google Scholar]
- O'Brien D. E.; Baiocchi F.; Robins R. K.; Cheng C. C. Pyrimidines. IX. 4- and 5-(Substituted-anilino)pyrimidines. J. Med. Pharm. Chem. 1962, 561085–1103. [DOI] [PubMed] [Google Scholar]
- Monaco H. L. Crystal structure of chicken riboflavin-binding protein. EMBO J. 1997, 16, 1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A. Interaction of riboflavin binding protein with riboflavin, quinacrine, chlorpromazine and daunomycin. Int. J. Biochem. 1988, 2091021–1029. [DOI] [PubMed] [Google Scholar]
- Choi J. D.; McCormick D. B. The interaction of flavins with egg white riboflavin-binding protein. Arch. Biochem. Biophys. 1980, 204141–51. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Sun Q.; Kini R. M.; Sivaraman J. A universal method for fishing target proteins from mixtures of biomolecules using isothermal titration calorimetry. Protein Sci. 2008, 17101798–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caelen I.; Kalman A.; Wahlstrom L. Biosensor-Based Determination of Riboflavin in Milk Samples. Anal. Chem. 2003, 761137–143. [DOI] [PubMed] [Google Scholar]
- Hong S.; Leroueil P. R.; Majoros I. J.; Orr B. G.; Baker J. J. R.; Banaszak Holl M. M. The Binding Avidity of a Nanoparticle-Based Multivalent Targeted Drug Delivery Platform. Chem. Biol. 2007, 141107–115. [DOI] [PubMed] [Google Scholar]
- Attie A. D.; Raines R. T. Analysis of Receptor-Ligand Interactions. J. Chem. Educ. 1995, 722119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavalitshewinkoon P.; Wilairat P.; Gamage S.; Denny W.; Figgitt D.; Ralph R. Structure-activity relationships and modes of action of 9-anilinoacridines against chloroquine-resistant Plasmodium falciparum in vitro. Antimicrob. Agents Chemother. 1993, 373403–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. J. The use of quinacrine (Atabrine) in rheumatic diseases: A reexamination. Semin. Arthritis Rheum. 1989, 184282–296. [DOI] [PubMed] [Google Scholar]
- Wilson W. D.; Jones R. L. Interaction of actinomycin D, ethidium quinacrine daunorubicin, and tetralysine with DNA: 31P NMR chemical shift and relaxation investigation. Nucleic Acids Res. 1982, 1041399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harle D. G.; Baldo B. A. Structural features of potent inhibitors of rat kidney histamine N-methyltransferase. Biochem. Pharmacol. 1988, 373385–388. [DOI] [PubMed] [Google Scholar]
- Stuhlmeier K. M.; Pollaschek C. Quinacrine but not chloroquine inhibits PMA induced upregulation of matrix metalloproteinases in leukocytes: quinacrine acts at the transcriptional level through a PLA2-independent mechanism. J. Rheumatol. 2006, 333472–480. [PubMed] [Google Scholar]
- Orzáez M.; Mondragón L.; García-Jareño A.; Mosulén S.; Pineda-Lucena A.; Pérez-Payá E. Deciphering the antitumoral activity of quinacrine: Binding to and inhibition of Bcl-xL. Bioorg. Med. Chem. Lett. 2009, 1961592–1595. [DOI] [PubMed] [Google Scholar]
- Dey A.; Tergaonkar V.; Lane D. P. Double-edged swords as cancer therapeutics: simultaneously targeting p53 and NF-[kappa]B pathways. Nat. Rev. Drug Discovery 2008, 7121031–1040. [DOI] [PubMed] [Google Scholar]
- Gurova K. V.; Hill J. E.; Guo C.; Prokvolit A.; Burdelya L. G.; Samoylova E.; Khodyakova A. V.; Ganapathi R.; Ganapathi M.; Tararova N. D.; Bosykh D.; Lvovskiy D.; Webb T. R.; Stark G. R.; Gudkov A. V. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kΒ-dependent mechanism of p53 suppression in tumors. Proc. Natl. Acad. Sci. U.S.A. 2005, 1024817448–17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen M.; Choi S. K.; Whitesides G. M. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem., Int. Ed. 1998, 37, 2755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.