Summary
The Notch signaling pathway is an evolutionarily-conserved intercellular signaling mechanism, and mutations in its components disrupt embryonic development in many organisms and cause inherited diseases in humans. The Jagged2 (Jag2) gene, which encodes a ligand for Notch pathway receptors, is required for craniofacial, limb and T cell development. Mice homozygous for a Jag2 null allele die at birth from cleft palate, precluding study of Jag2 function in postnatal and adult mice. We have generated a Jag2 conditional null allele by flanking the first two exons of the Jag2 gene with loxP sites. Cre-mediated deletion of the Jag2flox allele generates the Jag2del2 allele, which behaves genetically as a Jag2 null allele. This Jag2 conditional null allele will enable investigation of Jag2 function in a variety of tissue-specific contexts.
Keywords: Notch signaling, conditional null allele, Cre-loxP, gene targeting
The Notch signaling pathway is an evolutionarily conserved intercellular signaling mechanism. Mutations in Notch pathway components disrupt embryonic development in diverse multicellular organisms and cause cancers and inherited disease syndromes in humans (Bray, 2006; Fiuza and Arias, 2007). In mammals, genes of the Notch family (Notch1 through Notch4) encode Type 1 transmembrane protein receptors that interact with Type 1 transmembrane ligands encoded by genes of the Delta-like (Dll1, Dll3 and Dll4) and Jagged (Jag1 and Jag2) families. We have previously described the construction and analysis of a targeted null allele of the Jagged2 (Jag2) gene, termed Jag2del1 (originally named Jag2ΔDSL) (Jiang et al., 1998). Jag2del1/Jag2del1 homozygous mutant mice die at birth from completely penetrant cleft palate. The Jag2del1/Jag2del1 mice also exhibit soft tissue syndactyly, the fusion of the digits on the fore- and hindlimbs (Jiang et al., 1998). Cleft palate and soft tissue syndactyly are also exhibited at reduced penetrance by mice homozygous for the syndactylism mutation, a spontaneous Jag2 missense mutant allele (Jag2sm) that behaves genetically as a Jag2 hypomorphic allele (Casey et al., 2006; Sidow et al., 1997). To circumvent the neonatal lethality exhibited by Jag2del1/Jag2del1 null mutant mice and permit the study of Jag2 function in postnatal and adult mice, we describe here construction of an allele for conditional inactivation of Jag2 gene function using the Cre-loxP system.
The Jag2 gene spans approximately 21 kb on mouse Chromosome 12, and consists of 26 exons. To generate the Jag2floxneo targeting vector (Fig. 1a), a PGKneo selection cassette was introduced into intron 2. The PGKneo cassette was flanked by FRT sites for removal by Flpe recombinase (Farley et al., 2000), and by a loxP site distal to the PGKneo cassette in intron 2. A second loxP site was introduced approximately 1.1 kb upstream of the Jag2 translational start site. A diphtheria toxin cassette, at the terminus of the targeting vector, was introduced for negative selection against random integration of the targeting vector into the ES cell genome. The design of the Jag2floxneo allele permits removal of the PGKneo cassette by mating to a Flpe deleter mouse line (generating the Jag2flox allele), or removal of Jag2 genomic sequence (including promoter sequences and exons 1 and 2) and the PGKneo cassette by mating to a Cre deleter line. To distinguish the Jag2 mutant allele generated by Cre-mediated deletion of the Jag2flox or Jag2floxneo alleles from the Jag2del1 allele, our previously published Jag2 targeted null mutant allele (Jiang et al., 1998), we designate the Jag2 null allele generated by Cre recombinase-mediated deletion of the Jag2flox or Jag2floxneo alleles the Jag2del2 allele.
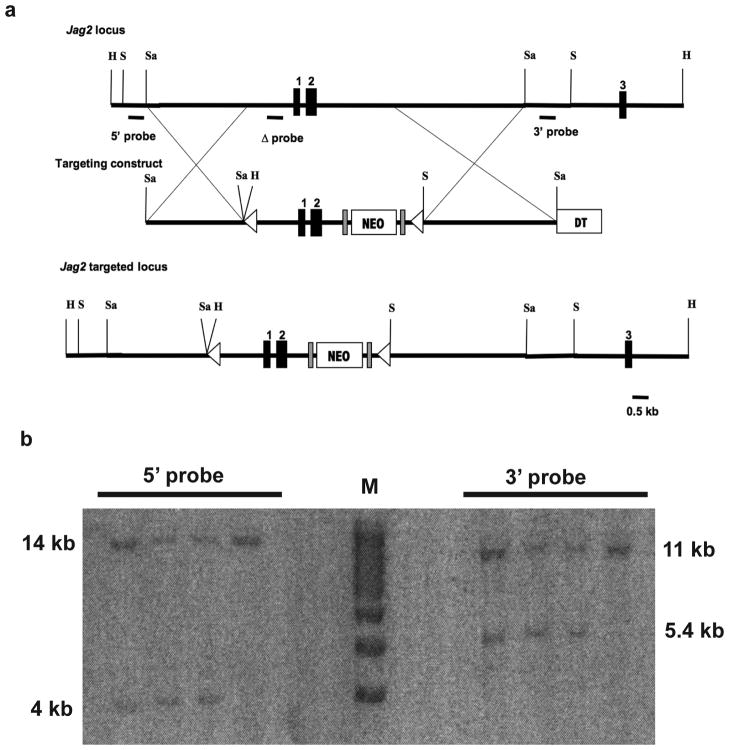
FIG. 1.
Generation of a Jag2 conditional null allele. (a) Schematic representation of a portion of the wildtype Jag2 allele, the Jag2floxneo targeting vector, and the targeted Jag2floxneo allele. Exons are indicated by boxes with coding sequences designated by dark shading. LoxP sequences are marked by triangles and FRT sites by gray rectangles. The FRT-PGKneo-FRT-LoxP- cassette was inserted between exons 2 and 3 and the second LoxP site was inserted 5′ to exon 1, approximately 1.1 kb upstream of the Jag2 translational start site. A diphtheria toxin (DT) cassette was included for negative selection of randomly integrated clones. The hybridization probe used for Southern blot analysis is indicated. Abbreviations: H, HindIII; S, ScaI; Sa, SacI. (b) Southern blot analysis of targeted ES cells. HindIII or ScaI digested ES cell genomic DNA was hybridized with the 5′ or 3′ probe respectively as indicated in a, yielding 14 kb wildtype and 4 kb mutant bands for the 5′ probe, and 11 kb wildtype and 5.4 kb mutant bands for the 3′ probe. M: markers.
The Jag2floxneo targeting construct was electroporated into R1 ES cells (Nagy et al., 1993), and three correctly targeted clones (Fig. 1b) were injected into C57BL/6J blastocysts. Chimeras were mated to C57BL/6J female mice, and germline transmission was obtained from one clone. Jag2floxneo/+ heterozygous mice were mated to a deleter line expressing the Flpe recombinase (Farley et al., 2000) to excise the PGKneo cassette and generate the Jag2flox allele. Mice heterozygous for the Jag2del2 null allele were generated by mating Jag2flox/+ heterozygous mice to Meox2-Cre mice which, in addition to other tissues, express Cre recombinase in the germline (Tallquist and Soriano, 2000). As with the Jag2del1 allele, heterozygous Jag2del2/+ mice were viable, fertile and displayed no obvious phenotypic abnormalities.
Cre-mediated excision of the Jag2flox allele to generate the Jag2del2 allele deletes predicted Jag2 promoter sequences, the ATG translation start site and exons 1 and 2, which we predict would create a Jag2 null allele. In order to assess the functionality of the Jag2del2 allele, embryos and neonatal mice homozygous for the Jag2del2 allele were obtained by intercrossing Jag2del2/+ heterozygous mice. We assessed Jag2 RNA expression in Jag2del2/Jag2del2 and Jag2del2/+ littermate control embryos at E10.5 by whole mount in situ hybridization. Jag2 RNA was not expressed at levels detectable by in situ hybridization in Jag2del2/Jag2del2 homozygous mutant embryos (Fig. 2). We also assessed Jag2 RNA expression in homozygous Jag2del2/Jag2del2 mice by quantitative RT-PCR. Using a primer set that spanned exons 3 and 4, Jag2del2/Jag2del2 homozygous mutants expressed 11.5 ± 1.3% of wildtype Jag2 transcript levels. Utilizing a primer set spanning exons 6–8 of the Jag2 gene, Jag2del2/Jag2del2 mutant embryos expressed 21.8± 3.6% of wildtype Jag2 transcript levels. However, these levels of the Jag2del2 mutant transcript do not appear to have any functional consequences, since Jag2del2/Jag2del2 homozygous mutant mice exhibited a phenotype identical to that exhibited by Jag2del1/Jag2del1 null mutant mice (Fig. 3). Jag2del2/Jag2del2 neonates died the first day of birth, exhibiting syndactyly of the fore- and hindlimbs and palate-tongue fusions that caused palatal clefting (Fig. 3). These data demonstrate that the Jag2del2 allele is a Jag2 null allele, functionally equivalent to the Jag2del1 allele, and that the extent of Cre-mediated deletion of the Jag2flox allele can be assessed by in situ hybridization. The neonatal lethality of Jag2 null mice has precluded analysis of Jag2 function in postnatal and adult mice. Utilization of the Jag2flox conditional null allele will enable the investigation of Jag2 function in a tissue-specific manner throughout the mouse life span.
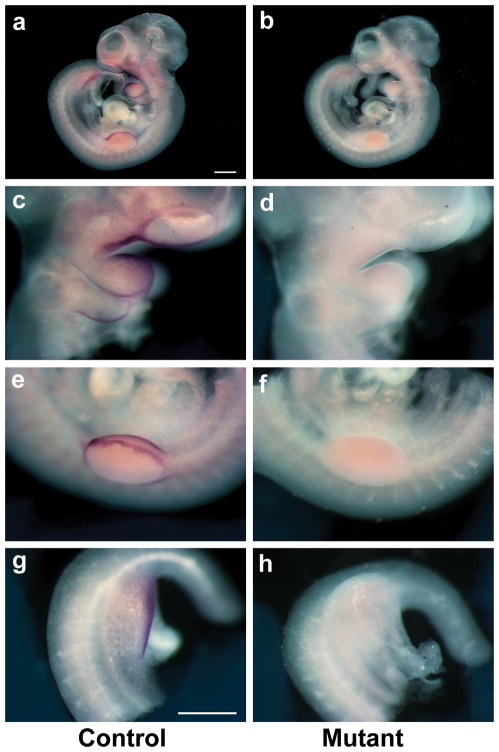
FIG. 2.
Absence of Jag2 RNA expression in Jag2del2/Jag2del2 embryos. (a,b) Whole mount in situ hybridization for Jag2 transcripts in Jag2del2/+ control littermate (a,c,e,g) and Jag2del2/Jag2del2 (b,d,f,h) embryos at E10.5. Jag2 RNA was not detectable by in situ hybridization in the mutant embryo. (c–h) Higher magnification views. (c,d) Branchial arches. (e,f) Apical ectodermal ridge of forelimb bud. (g,h) Hindlimb bud. Scale bars: a,b, 500 μm; c–g, 500 μm.
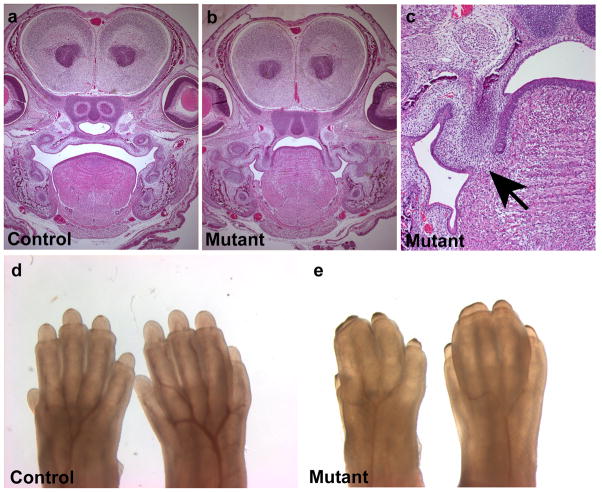
FIG. 3.
Phenotype of Jag2del2 homozygous mutant embryos. (a–c) Cleft palate in Jag2del2/Jag2del2 homozygotes. Coronal sections of E16.5 embryos show that the Jag2del2/Jag2del2 homozygote has cleft palate and fused tongue. (c) Fusion of the tongue with the unelevated palatal shelves in the Jag2del2/Jag2del2 homozygote (black arrow). (d,e) Limb defects in Jag2del2/Jag2del2 homozygotes. A forefoot (left) and hindfoot (right) of E16.5 mouse are shown in each panel. Jag2del2/Jag2del2 homozygous mutant embryos (e) exhibit syndactyly of both fore- and hindlimbs.
METHODS
Construction of the Jag2floxneo Allele
To construct the Jag2floxneo targeting vector, the 5′ LoxP site was cloned into the BclI site in the SacI digested fragment from the bMQ-136I11 strain 129 BAC clone, approximately 1.1 kb 5′ of the translational start site of the JAG2 protein. The polylinker sequences adjacent to the 5′ LoxP site introduce new SacI and HindIII sites, which are used for differentiation between the targeted and wildtype alleles by Southern blot analysis. An FRT-PGKneo-FRT-LoxP cassette with a ScaI site for Southern blot screening was inserted 3′ of exon 2 by bacterial recombineering (Warming et al., 2005). These fragments were assembled in a modified pBluescript II SK vector (Promega) containing a diphtheria toxin negative selection cassette.
Electroporation of ES cells and Generation of Jag2floxneo Mice
The Jag2floxneo targeting vector was linearized with SpeI and electroporated into R1 ES cells (Nagy et al., 1993). G418-resistant colonies were screened for homologous recombination by Southern blot hybridization. 10 μg of genomic DNA was digested with restriction enzymes, fractionated in 0.8% agarose gels, transferred to nylon membranes, and hybridized with P32-labelled probe. ES cells containing the expected recombination events were injected into C57BL/6J blastocysts. Male chimeric mice were bred with C57BL/6J females to obtain germ line transmission of the Jag2floxneo targeted allele.
Generation of the Jag2flox and Jag2del2 Alleles
Jag2floxneo/+ heterozygous mice were mated to a deleter line expressing the Flpe recombinase (Farley et al., 2000) to excise the PGKneo cassette and generate the Jag2flox allele. Excision of the PGKneo cassette was determined by PCR using the primers J2NeoF (CACCCCTCGAGCCCTTAAT) and J2NeoR (GCTGATCCGGAACCCTTAAT). Jag2flox/+ heterozygous mice were mated to the Meox2-Cre deleter line (Tallquist and Soriano, 2000) to excise Jag2 genomic sequences between the loxP sites and generate the Jag2del2 null allele. Excision of Jag2 genomic sequences was determined by PCR using the primers J2FlxF (GGCAGAGTAGCACATCACCA) and J2FlxR2 (ACATGCCTGCACAGCCTAC).
Mouse and Embryo Genotyping
Jag2floxneo mice were genotyped using 2 primers (J2FlxF, J2FlxR1: CAAGCGCACAGGTTGAGTAG) spanning the 5′ loxP site with a 401bp WT band and a 453 Flox band. Jag2flox mice were genotyped using primers spanning the deleted FNF cassette yielding a 202-bp mutant allele (J2NeoF and J2NeoR). The Jag2del2 allele was monitored using primers spanning the entire deleted region between the two LoxP sites, yielding a 302-bp band (J2FlxF and J2FlxR2). All PCR reactions were performed using the following standard cycling conditions: 94°C 3 min, (94°C 30 sec, 58°C 45 sec, 72°C 45 sec) X 39 cycles, 72°C 5 min.
Histological Analysis and In Situ Hybridization
We considered the day of vaginal plug as E0. E16.5 embryos were dissected and DNA was prepared from the tails for genotyping by PCR. Embryos were fixed in 10% formalin buffer. Feet were dissected and put into 30% glycerol for photography. For histological analysis, heads of the fixed embryos were dehydrated through graded alcohols, embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin. Whole mount in situ hybridization of E10.5 embryos was performed as described previously (Jiang et al., 1998). The Jag2 in situ hybridization probe was generated from a 1.4 kb partial Jag2 cDNA clone (Jiang et al., 1998), encoding EGF repeats 1–10 of the mouse JAG2 protein.
Reverse Transcription and Quantitative RT-PCR Analysis
For RT-PCR, total RNA was extracted from the E10.5 embryos, using the Trizol Plus RNA Purification System (Invitrogen, USA) and following the manufacturer’s instructions. 500 nanogram RNA was reverse-transcribed using the MessageSensor RT Kit (Ambion, USA). Two nanograms of cDNA were used for real-time PCR amplification for each well, using primers from Primerbank. All reactions were performed in triplicate using Power SYBR Green PCR Master Mix on a 7500 Real Time PCR system (Applied Biosystems). Data were normalized to Actb levels. Hey1 gene expression was used as an internal control. Primer sequences: Actb, F (forward): GGCTGTATTCCCCTCCATCG, R (reverse): CCAGTTGGTAACAATGCCATGT, product size 154 bp; Hey1, F: GCGCGGACGAGAATGGAAA, R: TCAGGTGATCCACAGTCATCTG, 231 bp; Jag2 Exons3–4, F: CAATGACACCACTCCAGATGAG, R: GGCCAAAGAAGTCGTTGCG, 203 bp; Jag2 Exons6–8, F: TGTGACGAGTGTGTCCCCTA, R: GTTCCATCCTGACGGACAGT, 318 bp.
Acknowledgments
We would like to thank Dr. Cara Bradley for helpful discussions and suggestions for generation of the targeting construct.
LITERATURE CITED
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Casey LM, Lan Y, Cho ES, Maltby KM, Gridley T, Jiang R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev Dyn. 2006;235:1830–1844. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidow A, Bulotsky MS, Kerrebrock AW, Bronson RT, Daly MJ, Reeve MP, Hawkins TL, Birren BW, Jaenisch R, Lander ES. Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature. 1997;389:722–725. doi: 10.1038/39587. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]