Abstract
Background
The prevalence of resistance to fusidic acid of methicillin-resistant Staphylococcus aureus (MRSA) was increased each year in a Taiwan hospital. Thirty-four MRSA clinical isolates collected in 2007 and 2008 with reduced susceptibility to FA were selected for further evaluation the presence of resistance determinants.
Results
The most common resistance determinant was fusC, found in 25 of the 34 MRSA isolates. One of the 25 fusidic acid-resistant MRSA harboured both fusB and fusC, which is the first time this has been identified. Mutations in fusA were found in 10 strains, a total of 3 amino-acid substitutions in EF-G (fusA gene) were detected. Two substitutions with G556S and R659L were identified for the first time. Low-level resistance to fusidic acid (MICs, ≤ 32 μg/ml) was found in most our collection. All collected isolates carried type III SCCmec elements. MLST showed the isolates were MRSA ST239. PFGE revealed nine different pulsotypes in one cluster.
Conclusions
Our results indicate that the increase in the number of fusidic acid resistant among the MRSA isolates in this hospital is due mainly to the distribution of fusC determinants. Moreover, more than one fusidic acid-resistance mechanism was first detected in a same stain in our collection.
Keywords: fusidic acid, MRSA, fusA, fusB, fusC
Background
The frequently-encountered multi-antibiotic resistance of MRSA has become a major health problem [1,2]. The prevalence of MRSA isolates, most of which are health care associated, has slowly increased since 1982, and the appearance and increasing incidence of community-associated MRSA infections has been documented. Globally, methicillin resistance among nosocomial S. aureus isolates is common [3,4].
Fusidic acid has been used to treat infections with S. aureus for over 35 years. It is usually used in combination with agents such as vancomycin or rifampin in the treatment of systemic infections caused by MRSA [5]. Fusidic acid inhibits protein synthesis by blocking the elongation of the nascent polypeptide chain through binding to EF-G on the ribosome and preventing the dissociation of EF-G⋅GDP from the ribosome [6,7]. The frequency of fusidic acid resistance is not very high; however, the emergence of clinical staphylococcal species that are resistant to fusidic acid has been reported [8-11].
The primary mechanism of fusidic acid resistance in S. aureus relates to mutations in fusA, the gene that encodes the ribosomal translocase and translation elongation factor EF-G [12,13]. More than 30 different amino acid substitution mutations in fusA have been identified [12,14,15]. Subsequently, resistance in natural isolates may also result from the horizontal acquisition of fusB, a poorly characterized plasmid-mediated resistance mechanism [13]. The gene fusB is usually carried by a 21-kb plasmid, pUB101 [16], however, it can also be chromosomal [17]. The fusB gene encodes an inducible protein that protects an in vitro translation system against the inhibitory action of fusidic acid [8]. Recently, two fusB homologues, designated fusC and fusD, have been identified in the chromosome of clinical isolates of S. aureus and S. saprophyticus, respectively [18]. In addition, fusidic acid-resistant small-colony variants (SCVs) of S. aureus with mutations in rplF have been designated as FusE mutants [14]. Although frequencies of resistance to fusidic acid have remained generally low, each of these mechanisms has multiple genetic causes, and emerging resistance is a problem that could limit the therapeutic options available for treatment of staphylococcal infections [19].
In this study, a series of MRSA clinical isolates recovered at a regional teaching hospital in middle Taiwan showing fusidic acid MIC ≥ 2 μg/ml. The high distribution of fusidic acid resistance determinants fusC was confirmed in MRSA. In addition, different fusidic acid resistance determinants-containing in one isolate was also demonstrated.
Methods
Bacterial isolates
From April 2007 to January 2008, 34 clinical isolates of MRSA with fusidic acid resistance were recovered from 34 different patients at Tungs' Taichung MetroHarbor Hospital (TTMHH), a 1405-bed regional teaching hospital in central Taiwan. S. aureus ATCC 29213 and NCTC 8325 have consistently been used as a quality control strain and Pulsed Field Gel Electrophoresis (PFGE) standard strain, respectively. Luria-Bertani (LB) agar and LB broth were used for bacterial growth at 37°C with aeration. Mueller-Hinton agar was used for all determinations of minimum inhibitory concentrations (MICs). All isolates were identified on the colony morphology, Gram's stain, a positive catalase reaction and/or results obtained with the phoenix system (BD Diagnostic Systems, Sparks, MD, USA) and frozen at -80°C until used.
Antimicrobial susceptibility tests
MICs of different antimicrobial agents were determined using the Phoenix Automated Microbiology System (BD Diagnostic Systems, Sparks, MD) and interpreted according to the criteria provided by the Clinical and Laboratory Standards Institute (CLSI). Fusidic acid susceptibility was screened by the disk diffusion method with 10 μg fusidic acid containing disks. The interpretive criterion of susceptibility was an inhibition zone ≥ 22 mm in diameter. Fusidic acid MICs were further determined by an agar dilution method following the CLSI guidelines, and susceptibility was categorized using the European Committee for Antimicrobial Susceptibility Testing (EUCAST)/British Society of Antimicrobial Chemotherapy (BSAC) criteria (susceptible, MIC < 2 μg/ml; resistant, MIC ≥ 2 μg/ml). The testing MIC range of fusidic acid was 0.12-512 μg/ml.
DNA manipulation and PCR
Total DNA from three to five isolated colonies was prepared using a Wizard genomic DNA preparation kit (Promega, Madison, WI) with 0.5 mg/ml of lysostaphin and 0.3 mg/ml of RNase for the lysis step. The multiplex PCR assay for fusB and fusC used oligonucleotide primers BF (5'-CTATAATGATATTAATGAGATTTTTGG), BR (5'-TTTTTACATATTGACCATCCGAATTGG), CF (5'-TTAAAGAAAAAGATATTGATATCTCGG), and CR (5'-TTTACAGAATCCTTTTACTTTATTTGG) to generate amplicons of 431 and 332 bp from the fusB and fusC genes, respectively. The cycling conditions consisted of an initial denaturation step (94°C for 3 min), followed by 25 cycles of 94°C (30 s), 57°C (30 s) and 72°C (45 s) [20]. For further identification of the fusB and fusC genes, primers FusB-R (5'-ACAGGATCCATTTTCACAAACATAGT) and FusB-F1(5'-AGGGATCCCATATTTAAAGCTATTG) were used to generate an amplicon comprising the 642 bp fusB with 122 bp of upstream DNA [8], and primers sas0043U (5'-GTAGGATCCATTGGGAATGATAAATAGTGA) and sas0043L (5'-TTTGGATCCATCGATTAAGAGTGAGGTACA) were used to generate a 2.5 kb amplicon with fusC [18]. The fusA gene was PCR-amplified using oligonucleotide primers rpsU and tufL and sequenced with these and three additional primers (AintS1, 5'-TAAGGGTCAGTCATAACTTT; AintS2, 5'-TTCAAAAACAAAGGTGTTCA; and AintS3, 5'-ATGTATTCACGAGGAAC) [20]. The PCR products were electrophoresed in 1.5% agarose gels and visualized under ultraviolet light. The PCR products were then purified with a commercial kit and both strands of the amplicons were sequenced on an ABI PRISM 370 automated sequencer (PE Applied Biosystems, Franklin Lakes, NJ). Sequence analyses were performed online at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Southern blot hybridization
DNA samples were digested by EcoR1 and analyzed by electrophoresis at 30 V for 2 h in a 1% w/v agarose gel. The gel was denatured in a solution of 0.5 M NaOH and 1.5 M NaCl, neutralized in 0.5 M Tris-HCl (pH 7.5) and 1.5 M NaCl on Whatman filter paper (Maidstone, UK), and finally saturated with 10% w/v SDS (15 min for each step). DNA was transferred to a positively charged nylon membrane (Boehringer Mannheim, Mannheim, Germany) using an electrophoretic transfer cell (Bio-Rad Laboratories, Hercules, CA). A probe for fusC was prepared by randomly labelling the 2.5 kb PCR product of fusC with digoxigenin using a commercial kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. The fusC gene for the hybridization probe was amplified using oligonucleotide primers fusCU 5'-GAGGAATATCATATGAATAAAATAGAAGTGTA and fusCL 5'-AGAGTGGATCCCAAAATATAACAACCCTGATC [18].
SCCmec typing by PCR
The presence of mecA was determined using the primers MR1 5'-GTGGAATTGGCCAATACAGG and MR2 5'-TGAGTTCTGCAGTACCGGAT, which were used to PCR-amplify a 1,339 bp internal fragment of the gene [21]. PCR was carried out for 30 cycles of 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C. Characterization of SCCmec elements was performed by multiple PCR as previously described [22].
PFGE and multilocus sequence typing (MLST)
Genotyping of S. aureus strains was conducted by macrorestriction of bacterial DNA followed by PFGE separation of the resulting fragments. Whole chromosomal DNA of the clinical isolates embedded in agarose gel plugs (FMC Bioproducts, Philadelphia, PA) were treated with proteinase K and SmaI restriction endonuclease according to the manufacturer's recommendations (New England Biolabs, Ipswich, MA). PFGE and DNA fingerprints analysis were performed as described previously [23]. The isolates were also analyzed by MLST as described previously [24].
Plasmid curing
The clinical isolate with pUB101-like plasmid was subjected to elevated temperature-mediated plasmid elimination by sequential passages in LB (approximately 100 cells into 100 ml) at 43°C with shaking for about 30 generations. Cured strains were diluted and plated on LA plates (LB plus 1% agar; Merck, Darmstadt, Germany) to obtain single colonies. Loss of cadmium resistance was screened by replica plating at 37°C [25]. Loss of the plasmid was confirmed by loss of unselected phenotypic traits (ampicillin resistance) and by PCR of cadXD [15].
Ethics
This study was reviewed by the Institutional Review Board (IRB) of the TTMHH and it was decided not to constitute the research involving human subject. An exemption certificate was issued by the IRB to attest this fact.
Results
Isolates and susceptibility tests
The sources of the 34 fusidic acid-resistant MRSA isolates included sputum (n = 9), pus (n = 16), blood (n = 5), urine (n = 2), ascites (n = 1), and tip of a central venous catheter (n = 1) (Table 1). All 34 clinical isolates were analyzed in more detail with regard to their antibiotic resistance profiles, and they were all susceptible to vancomycin, teicoplanin, quinupristin-dalfopristin, linezolid, and nitrofurantoin. The MICs for fusidic acid (2-64 μg/ml) were low to moderate level resistance phenotype. All isolates were uniformly resistant to penicillin, ampicillin, oxacillin, clindamycin, erythromycin, ciprofloxacin and gentamicin. The susceptible rates and MIC ranges of other antibiotics were as follows: rifampin 91%; chloramphenicol 88%; moxifloxacin 6%; levofloxacin 3%; tetracycline 3%; and trimethoprim-sulfamethoxazole 3%. The study results revealed that fusidic acid-resistant S. aureus was resistant to nearly all tested antibiotics except for vancomycin, teicoplanin, linezolid, nitrofurantoin, quinupristin-dalfopristin, chloramphenicol, and rifampin.
Table 1.
Characteristics and mechanisms of the 34 fusidic acid-resistant MRSA clinical isolates
| Isolate | Specimen | FA MIC (μg/ml) |
VAN MIC (μg/ml) |
LZD MIC (μg/ml) |
OXA MIC (μg/ml) |
RIF MIC (μg/ml) |
fusC |
Polymorphism(s) in EF-G |
PFGE patterns |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Pus | 8 | 1 | 1 | >2 | < = 0.5 | - | H457Y | A1 |
| 2 | Pus | 32 | 1 | 2 | >2 | < = 0.5 | + | - | A1 |
| 3 | Pus | 8 | 1 | 1 | >2 | < = 0.5 | + | - | A1 |
| 4 | Sputum | 16 | 1 | 2 | >2 | < = 0.5 | + | H457Y | A1 |
| 5 | Sputum | 32 | 2 | 2 | >2 | >2 | + | - | A1 |
| 6 | Pus | 16 | 1 | 1 | >2 | >2 | + | - | A2 |
| 7 | Pus | 8 | 1 | 1 | >2 | < = 0.5 | + | - | A3 |
| 8 | Sputum | 16 | 1 | 1 | >2 | < = 0.5 | + | - | A3 |
| 9 | Pus | 16 | 1 | 1 | >2 | < = 0.5 | - | G556S | A3 |
| 10 | Sputum | 16 | 1 | 1 | >2 | < = 0.5 | - | H457Y, G556S | A3 |
| 11 | Ascites | 8 | 1 | 1 | >2 | < = 0.5 | - | H457Y | A3 |
| 12 | Pus | 64 | 2 | 2 | >2 | < = 0.5 | + | - | A3 |
| 13 | Sputum | 64 | 2 | 2 | >2 | < = 0.5 | - | H457Y | A3 |
| 14 | Pus | 16 | 1 | 1 | >2 | < = 0.5 | + | - | A3 |
| 15 | Blood | 4 | 1 | 1 | >2 | < = 0.5 | + | - | A3 |
| 16 | Pus | 8 | 1 | 1 | >2 | < = 0.5 | + | - | A3 |
| 17 | Blood | 8 | 1 | 1 | >2 | < = 0.5 | + | - | A3 |
| 18 | Blood | 16 | 1 | 1 | >2 | < = 0.5 | + | - | A3 |
| 19 | Blood | 16 | 1 | 1 | >2 | < = 0.5 | + | - | A3 |
| 20 | Pus | 2 | 2 | 1 | >2 | < = 0.5 | + | - | A3 |
| 21 | Urine | 2 | 2 | 2 | >2 | < = 0.5 | - | H457Y, G556S | A3 |
| 22 | Sputum | 2 | 2 | 2 | >2 | < = 0.5 | + | - | A3 |
| 23 | Pus | 16 | 2 | 1 | >2 | >2 | - | H457Y | A4 |
| 24 | Pus | 2 | 1 | 1 | >2 | < = 0.5 | + | - | A5 |
| 25 | Urine | 16 | 1 | 1 | >2 | < = 0.5 | + | - | A6 |
| 26 | CVP tip | 8 | 1 | 2 | >2 | < = 0.5 | + | - | A6 |
| 27 | Pus | 2 | 2 | 2 | >2 | < = 0.5 | + | - | A6 |
| 28 | Sputum | 16 | 1 | 2 | >2 | < = 0.5 | + | - | A7 |
| 29a | Pus | 8 | 1 | 2 | >2 | < = 0.5 | + | - | A8 |
| 30 | Sputum | 16 | 1 | 2 | >2 | < = 0.5 | + | - | A9 |
| 31 | Pus | 16 | 1 | 2 | >2 | < = 0.5 | - | H457Y, R659L | A9 |
| 32 | Sputum | 8 | 1 | 2 | >2 | < = 0.5 | + | - | A9 |
| 33 | Blood | 16 | 1 | 1 | >2 | < = 0.5 | - | G556S | A9 |
| 34 | Pus | 2 | 2 | 2 | >2 | < = 0.5 | + | - | A9 |
FA, fusidic acid; VAN, vancomycin; LZD, linezolid; OXA, oxacillin; RIF, rifampin
a nonsense mutation in fusC (S175 was encoded by TAA rather than TCA)
Genetic basis of resistance to fusidic acid: fusB and fusC
The genetic basis for resistance to fusidic acid in the isolates was determined by a multiplex PCR assay capable of detecting both the 431 bp fusB and 332 bp fusC genes [20]. Twenty-five of the 34 isolates (73.5%) were found to harbour the gene encoding fusC and one (isolate 32) among the 25 isolates also harboured the gene encoding fusB. Furthermore, using plasmid DNA of isolate 32 as a template, PCR with FusB-specific primers FusB-R1 and FusB-F1 and subsequent sequence analysis of the 764 bp PCR product confirmed the 100% identity of the fusB gene from plasmid pUB101. A curing study revealed that both the cadXD and fusB genes were plasmid encoded, and that fusC remained in the plasmid cured isolate 32. The MIC of fusidic acid for isolate 32 was 8 μg/ml after curing of the plasmid.
The full-length fusC gene was identified by PCR and sequenced in isolates 4, 24, 29, 30, and 32. The alignment of the amino acid sequences deduced from these isolates 4, 24, 30, and 32 fusC DNA sequences revealed 100% identity with FusC protein of S. aureus MSSA476 [18]. However, fusC from isolate 29 carried a nonsense mutation (S175 was encoded by TAA rather than TCA) that produced a change from fusidic acid resistance (MIC = 8 μg/ml) to fusidic acid susceptibility (MIC < 0.125 μg/ml) following two non-selective subcultures. The other isolates were screened for the presence of the fusC gene by Southern hybridization, and all tested isolates were positive for fusC (Figure 1).
Figure 1.
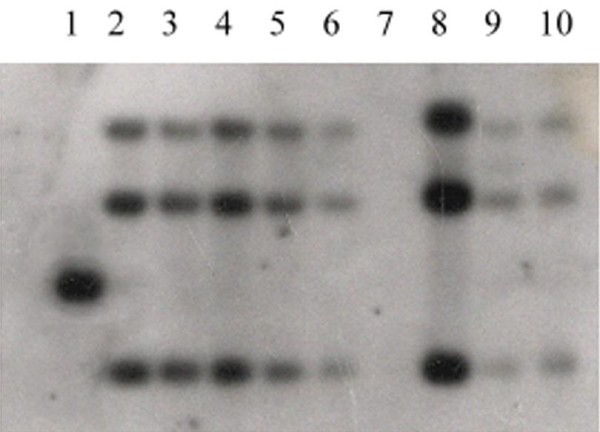
Southern hybridization of fusC. Detection of fusC by Southern hybridization in eight representatives of clinical fusidic acid-resistant S. aureus isolates that did not harbour fusB or resistance polymorphisms in fusA. Lane 1: 2.5-kb PCR fusC fragment from strain 2 as the positive control. Lanes 2-6 and 8-10: strains 3, 6, 15, 18, 24, 28, 29 and 34, respectively. Lane 7: strain 23 without the fusC gene. All total DNA was EcoRI-digested.
Detection of fusA gene mutations
PCR amplification and complete sequencing were performed to detect fusA gene mutations in the 34 isolates (Table 1). Five isolates possessed a mutation in H457Y, two isolates (isolates 9 and 33) exhibited a G556S mutation, and two isolates (isolates 10 and 21) harboured mutations in H457Y and G556S. In addition, isolate 31 possessed a mutation in H457Y and R659L. Single amino acid substitutions were found in seven isolates, and two amino acid substitutions were found in the other three. This is the first time that two different amino acid substitutions, G556S and R659L, have been reported in fusA gene mutations. Furthermore, one isolate (isolate 4) was encoded with fusC and fusA gene mutation. In this study, the most common amino acid substitution H457Y did not result in a high level of fusidic acid resistance (MIC ≥ 128 μg/ml).
Molecular epidemiological analysis
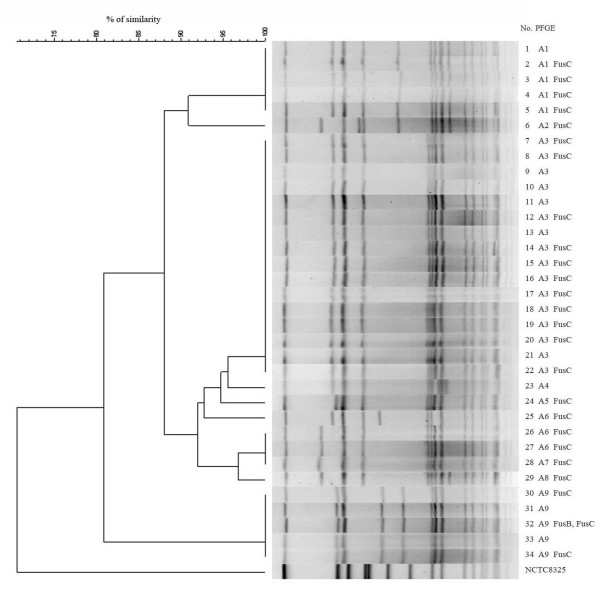
All 34 isolates included in this study met the criteria of being health care associated. The genotype analyses and their frequencies are shown in Table 1. Only one defined MLST type (ST239) was evident. All 34 isolates carried SCCmec type III elements. PFGE patterns of SmaI macrorestriction fragment analysis of these 34 isolates revealed nine distinct pulsotypes (A1-A9) that were classified into one cluster (> 80% similarity) (Figure 2). The results of PFGE patterns are summarized in Table 1.
Figure 2.
SmaI PFGE patterns of the 34 clinical fusidic acid-resistant Staphylococcus aureus isolates. PFGE patterns analysis of these 34 isolates revealed nine distinct pulsotypes (A1-A9) that were classified into one cluster.
Discussion
Previous studies of fusidic acid-resistance in clinical isolates have mostly focused on methicillin-susceptible S. aureus (MSSA) and other staphylococci [17,20,26]. Chen et al. recently reported that the prevalence of fusidic acid-resistance determinants was quite different between MRSA and MSSA groups [27]. In northern Taiwan collections, the fusA mutations were the major determinant (84%) followed by fusC with 16% fusidic acid-resistance in MRSA isolates [27]. In the present study based in central Taiwan, we found that the fusidic acid-resistant predominant determinant in MRSA was a high prevalence of fusC with 74% in clinical isolates. Furthermore, one isolate carried the fusB determinant on the plasmid and fusC determinant on the chromosome in a clinical fusidic acid-resistant S. aureus isolate. The FusC protein has a 45% amino acid similarity to FusB. The fusC gene was originally identified in the genome sequence of S. aureus MSSA476, and has been reported in fusidic acid-resistant S. intermedius and S. epidermidis [18,20]. In most European collections, fusC has been shown to be responsible for resistance to fusidic acid in all S. aureus strains examined that do not carry fusB or resistance mutations in fusA [17,18]. Moreover, the fusB gene has only been detected in MSSA, not in MRSA in most clinical collections in Taiwan [27]. Therefore, the present study shows the spread of fusC in Taiwan and for the first time demonstrates the presence of both fusB and fusC in a MRSA clinical isolate.
The most common mutation in fusA that conferred resistance to fusidic acid was the substitution H457Y in our study (Table 1). We reviewed the English literature and did not find any reports of two amino acid substitutions in EF-G of G556S and R659L relative to the resistance of fusidic acid. Mutations in EF-G are associated with fitness cost in the fusidic acid-resistance of S. aureus in vitro and in vivo [12,14]. The resistance mutations with amino acid substitutions occur mostly in structural domain III of EF-G, but some occur in domains I and V [28,29]. We identified a novel substitution present in fusidic acid-resistant S. aureus (isolates 9 and 33), which conferred an identical resistance mutation in fusA (G556S). The two isolates exhibited resistance to fusidic acid with MIC = 16 μg/ml and carried neither fusB nor fusC. In addition, substitution G556S was found in isolates 10 and 21 and was accompanied by mutations in fusA (H457Y). Another novel substitution amino acid substitution R659L located in domain V of EF-G was found to be accompanied with fusC mutations in our study. The role of this newly found amino acid substitution in fusA on the level of resistance is unknown and needs further investigation. Of the 34 isolates that were studied completely, isolate 4 harboured fusC and a resistance mutation in fusA (H457Y). This indicates that the fusidic acid-resistance in these MRSA clinical isolates had multiple genetic lineages.
The isolates with fusB and fusC determinants usually displayed higher level resistance to fusidic acid (> 16 μg/ml) [8,17]. The MICs of fusidic acid in our collections carrying fusC ranged from 2-64 μg/ml. It is not clear the reason why in non-selective subcultures, isolate 29 with one mutation site of the fusC gene lost the resistance to fusidic acid. We hypothesized that the mutation may result in FusC truncated after amino acid 174, and thus isolate 29 became susceptible. In this study, the single-amino-acid substitutions in EF-G substitution did not result in a high level fusidic acid resistance which is similar to previous report in MRSA strains belonging to CC8, H457Y mutation was associated with MIC of 64 μg/L and H457Q was associated with MIC of 4 μg/L [30]. The level of fusidic acid resistance in the isolate 4 with two fusidic acid resistance determinants couldn't be accounted for by their genotypes when compared with other clinical isolates with one of the determinants. A previous study showed a similar result that a laboratory strain containing both fusA resistance mutation and fusB failed to increase the level of fusidic acid resistance [17]. The chromosomal gene fusC confer resistance to fusidic acid on S. aureus or S. intermedius is identified with 45% amino acid similarity to FusB, protect EF-G from the antibiotic [18]. Genes for FusB-type resistance (fusB and fusC) are thought to act by the same mechanism of protection the drug target [18]. It remains unclear whether these resistance mechanisms of a strain do act in combination or not. The precise action mode of FusB-type resistance awaits further investigation. The level of fusidic acid resistance in isolate 32 did not decrease after curing the pUB101 plasmid. The result may indicate that the resistance mechanisms do not act synergistically or additively.
In this study, all MRSA isolates met the criteria of being health-care associated. PFGE patterns revealed that there was greater than 80% similarity among the isolates. MLST and SCCmec typing showed that all isolates belonged to ST239 and carried SCCmec III elements, which is the most prevalent health care-associated strain of MRSA in Taiwan [31]. A previous study conducted in 2002-2007 in northern Taiwan also revealed that most of fusidic acid-resistant MRSA isolates carried SCCmec type III [27]. The two studies results suggest that a clonal strain had disseminated in Taiwan during the period of the study. In contrast to our findings, a previous European study finding indicated that the majority of fusidic acid-resistant MRSA isolates belonged to CC80-MRSA-IV clone carrying fusB and CC5 clone harbouring fusC [30].
Conclusion
In conclusion, we hypothesize that the prevalence of fusidic acid-resistance in S. aureus was commonly associated with the fusC determinant in our isolates. It is interesting to note that some studied isolates possessed more than one fusidic acid-resistance mechanism in our collection. The fusC and acquired FusB-family determinants in a single isolate were first detected and one isolate with fusC also carried a fusA mutation in H457Y. Phylogenetic analysis clearly demonstrated the spread of a major clonal strain of fusidic acid-resistant MRSA in our institution. Due to the concern of clonal spread and growing expansion of fusidic acid-resistant determinants, particularly FusC in MRSA, large-scale, prospective surveillance monitoring for fusidic acid-resistance in S. aureus and MRSA is now ongoing in Taiwan.
Abbreviations
BSAC: British Society of Antimicrobial Chemotherapy; CLSI: Clinical and Laboratory Standards Institute; EUCAST: European Committee for Antimicrobial Susceptibility Testing; FA: fusidic acid; IRB: Institutional Review Board; LB: Luria-Bertani; MICs: minimum inhibitory concentrations; MLST: multilocus sequence typing; MRSA: methicillin-resistant Staphylococcus aureus; MSSA: methicillin-susceptible S. aureus; PCR: polymerase chain reaction; PFGE: pulsed field gel electrophoresis; SCC: staphylococcal chromosome cassette; SCVs: small-colony variants; TTMHH: Tungs' Taichung MetroHarbor Hospital.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CMC planned the idea and prepared the manuscript. MH participated in the study design and provided resources of experimental work. HFC conducted the experimental work. SCK and CRL provided technical help with PFGE and MLST. JHW supervised study design. LTW conceived this study, participated in its design, and the coordination and writing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Chih-Ming Chen, Email: chioming2002@yahoo.com.tw.
Mei Huang, Email: meihuang99@gmail.com.
Huei-Fen Chen, Email: ada741022@yahoo.com.tw.
Se-Chin Ke, Email: t0277@ms.sltung.com.tw.
Chia-Ru Li, Email: t6394@ms.sltung.com.tw.
Jen-Hsien Wang, Email: jenhsien@www.cmuh.org.tw.
Lii-Tzu Wu, Email: ltwu@mail.cmu.edu.tw.
Acknowledgements
We wish to thank Chien-Shun Chiou of the third branch office of Centers for Diseases Control of Taiwan for his assistance in PFGE analysis. This work was supported in part by research grant CMU97-104 from the China Medical University.
References
- Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10(3):505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM. Antibiotic resistance in staphylococci. Int J Antimicrob Agents. 2000;16(Suppl 1):S3–10. doi: 10.1016/s0924-8579(00)00299-5. [DOI] [PubMed] [Google Scholar]
- Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368(9538):874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- Gould SW, Rollason J, Hilton AC, Cuschieri P, McAuliffe L, Easmon SL, Fielder MD. UK epidemic strains of meticillin-resistant Staphylococcus aureus in clinical samples from Malta. J Med Microbiol. 2008;57(Pt 11):1394–1398. doi: 10.1099/jmm.0.2008/003509-0. [DOI] [PubMed] [Google Scholar]
- Whitby M. Fusidic acid in the treatment of methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 1999;12(Suppl 2):S67–71. doi: 10.1016/s0924-8579(98)00075-2. [DOI] [PubMed] [Google Scholar]
- Bodley JW, Zieve FJ, Lin L, Zieve ST. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem Biophys Res Commun. 1969;37(3):437–443. doi: 10.1016/0006-291X(69)90934-6. [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326(5953):694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill AJ, Chopra I. Molecular basis of fusB-mediated resistance to fusidic acid in Staphylococcus aureus. Mol Microbiol. 2006;59(2):664–676. doi: 10.1111/j.1365-2958.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- O'Neill AJ, Larsen AR, Skov R, Henriksen AS, Chopra I. Characterization of the epidemic European fusidic acid-resistant impetigo clone of Staphylococcus aureus. J Clin Microbiol. 2007;45(5):1505–1510. doi: 10.1128/JCM.01984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N, Afzal-Shah M, Warner M, Livermore DM. In vitro activity of retapamulin against Staphylococcus aureus isolates resistant to fusidic acid and mupirocin. J Antimicrob Chemother. 2008;62(4):766–768. doi: 10.1093/jac/dkn266. [DOI] [PubMed] [Google Scholar]
- Osterlund A, Kahlmeter G, Haeggman S, Olsson-Liljequist B. Staphylococcus aureus resistant to fusidic acid among Swedish children: a follow-up study. Scand J Infect Dis. 2006;38(5):334–334. doi: 10.1080/00365540500491976. [DOI] [PubMed] [Google Scholar]
- Nagaev I, Bjorkman J, Andersson DI, Hughes D. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol Microbiol. 2001;40(2):433–439. doi: 10.1046/j.1365-2958.2001.02389.x. [DOI] [PubMed] [Google Scholar]
- Turnidge J, Collignon P. Resistance to fusidic acid. Int J Antimicrob Agents. 1999;12(Suppl 2):S35–44. doi: 10.1016/s0924-8579(98)00072-7. [DOI] [PubMed] [Google Scholar]
- Norstrom T, Lannergard J, Hughes D. Genetic and phenotypic identification of fusidic acid-resistant mutants with the small-colony-variant phenotype in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(12):4438–4446. doi: 10.1128/AAC.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannergard J, Norstrom T, Hughes D. Genetic determinants of resistance to fusidic acid among clinical bacteremia isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(5):2059–2065. doi: 10.1128/AAC.00871-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien FG, Price C, Grubb WB, Gustafson JE. Genetic characterization of the fusidic acid and cadmium resistance determinants of Staphylococcus aureus plasmid pUB101. J Antimicrob Chemother. 2002;50(3):313–321. doi: 10.1093/jac/dkf153. [DOI] [PubMed] [Google Scholar]
- O'Neill AJ, Larsen AR, Henriksen AS, Chopra I. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob Agents Chemother. 2004;48(9):3594–3597. doi: 10.1128/AAC.48.9.3594-3597.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill AJ, McLaws F, Kahlmeter G, Henriksen AS, Chopra I. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob Agents Chemother. 2007;51(5):1737–1740. doi: 10.1128/AAC.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie D, Gray J. Fusidic acid resistance in Staphylococcus aureus. Arch Dis Child. 2004;89(1):74–77. doi: 10.1136/adc.2003.019695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaws F, Chopra I, O'Neill AJ. High prevalence of resistance to fusidic acid in clinical isolates of Staphylococcus epidermidis. J Antimicrob Chemother. 2008;61(5):1040–1043. doi: 10.1093/jac/dkn071. [DOI] [PubMed] [Google Scholar]
- Weller TM. The distribution of mecA, mecR1 and mecI and sequence analysis of mecI and the mec promoter region in staphylococci expressing resistance to methicillin. J Antimicrob Chemother. 1999;43(1):15–22. doi: 10.1093/jac/43.1.15. [DOI] [PubMed] [Google Scholar]
- Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41(11):5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey RW, Grinsted J. Linkage of fusidic acid resistance to the penicillinase plasmid in Staphylococcus aureus. J Gen Microbiol. 1972;73(3):501–508. doi: 10.1099/00221287-73-3-501. [DOI] [PubMed] [Google Scholar]
- Osterlund A, Eden T, Olsson-Liljequist B, Haeggman S, Kahlmeter G. Clonal spread among Swedish children of a Staphylococcus aureus strain resistant to fusidic acid. Scand J Infect Dis. 2002;34(10):729–734. doi: 10.1080/0036554021000026926. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Hung WC, Tseng SP, Tsai JC, Hsueh PR, Teng LJ. Fusidic Acid Resistance Determinants in Staphylococcus aureus Clinical Isolates. Antimicrob Agents Chemother. 2010;54(12):4985–4991. doi: 10.1128/AAC.00523-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Kristensen O, Martemyanov K, Gudkov AT, Nagaev I, Hughes D, Liljas A. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J Mol Biol. 2000;303(4):593–603. doi: 10.1006/jmbi.2000.4168. [DOI] [PubMed] [Google Scholar]
- Chen Y, Koripella RK, Sanyal S, Selmer M. Staphylococcus aureus elongation factor G--structure and analysis of a target for fusidic acid. FEBS J. 2010;277(18):3789–3803. doi: 10.1111/j.1742-4658.2010.07780.x. [DOI] [PubMed] [Google Scholar]
- McLaws FB, Larsen AR, Skov RL, Chopra I, O'Neill AJ. Distribution of Fusidic Acid Resistance Determinants in Methicillin-Resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55(3):1173–1176. doi: 10.1128/AAC.00817-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Tseng SP, Hu JM, Tsai JC, Hsueh PR, Teng LJ. Clonal spread of SCCmec type IV methicillin-resistant Staphylococcus aureus between community and hospital. Clin Microbiol Infect. 2007;13(7):717–724. doi: 10.1111/j.1469-0691.2007.01718.x. [DOI] [PubMed] [Google Scholar]