Abstract
Ral is a small Ras-like GTPase that regulates membrane trafficking and signaling. Here, we show that in response to planar cell polarity (PCP) signals, Ral modulates asymmetric Notch signaling in the Drosophila eye. Specification of the initially equivalent R3/R4 photoreceptor precursor cells in each developing ommatidium occurs in response to a gradient of Frizzled (Fz) signaling. The cell with the most Fz signal (R3) activates the Notch receptor in the adjacent cell (R4) via the ligand Delta, resulting in R3/R4 cell determination and their asymmetric positions within the ommatidium. Two mechanisms have been proposed for ensuring that the cell with the most Fz activation sends the Delta signal: Fz-dependent transcriptional upregulation in R3 of genes that promote Delta signaling, and direct blockage of Notch receptor activation in R3 by localization of an activated Fz/Disheveled protein complex to the side of the plasma membrane adjacent to R4. Here, we discover a distinct mechanism for biasing the direction of Notch signaling that depends on Ral. Using genetic experiments in vivo, we show that, in direct response to Fz signaling, Ral transcription is upregulated in R3, and Ral represses ligand-independent activation of Notch in R3. Thus, prevention of ligand-independent Notch activation is not simply a constitutive process, but is a target for regulation by Ral during cell fate specification and pattern formation.
Keywords: Ral GTPase (Rala), Notch/Delta signaling, Drosophila eye, Planar cell polarity
INTRODUCTION
Functions for Ral (Rala – FlyBase), a small Ras-like GTPase, are only beginning to be discovered. Ral has a well-characterized role in secretion (Moskalenko et al., 2001; Sugihara et al., 2002), and is also implicated in other membrane trafficking and remodeling events (Feig, 2003; van Dam and Robinson, 2006; Chen et al., 2006; Cascone et al., 2008; Wu et al., 2008; Spiczka and Yeaman, 2008; Lalli, 2009; Hase et al., 2009). Ral also regulates Rheb-dependent nutrient sensing in vertebrate cells (Maehama et al., 2008), Jak/Stat- and JNK-dependent apoptotic pathways in Drosophila (Balakireva et al., 2006; Ghiglione et al., 2008), and vertebrate tumor cell survival (Camonis and White, 2005; Chien et al., 2006). Here, we describe a specific role for Ral in PCP-dependent Notch signaling that patterns the Drosophila eye.
The Drosophila eye exhibits PCP in the arrangement of its ommatidia, or facets (Wolff and Ready, 1993). There are two chiral forms of ommatidia, dorsal and ventral, reflected through the dorsal/ventral midline, or equator. Ommatidial polarity is governed by the Fz/PCP signaling pathway, which has common core components in vertebrates and Drosophila (Strutt and Strutt, 2005; Klein and Mlodzik, 2005; Lawrence et al., 2007; Strutt and Strutt, 2009; Wu and Mlodzik, 2009; Axelrod, 2009; Simons and Mlodzik, 2008). Ommatidial chirality is defined by a pair of photoreceptors, R3 and R4, at the apex of a trapezoidal arrangement of eight photoreceptors. The presumptive R3 is closer to the equator, and thus, early in eye development, has higher levels of Fz activation than the presumptive R4. Fz asymmetry results in the pre-R3 cell, via the ligand Delta, activating the Notch receptor in the pre-R4 cell (Fanto and Mlodzik, 1999; Cooper and Bray, 1999; Tomlinson and Struhl, 1999). Asymmetric Notch activation in the R3/R4 pair ultimately determines the chirality of the ommatidium. Two different mechanisms, which are not necessarily mutually exclusive, have been proposed to explain how the difference in Fz activation leads to asymmetric Delta/Notch signaling. In one model, elevated Fz activation in pre-R3 leads directly to elevated transcription of Delta and neuralized (neur), which promote Delta signaling in R3 (Fanto and Mlodzik, 1999; del Alamo and Mlodzik, 2006). neur encodes a ubiquitin ligase that ubiquitylates Delta, and is required for Delta signaling. Subsequently, Notch activation in pre-R4 suppresses Delta and neur expression, which leads to less Notch activation in pre-R3 via a feedback loop. Alternatively (or in addition), Fz activation polarizes cells by localizing a Fz/Disheveled complex to one side of the plasma membrane (Strutt et al., 2002) (see also Tomlinson and Struhl, 1999). Disheveled (Dsh) is a cortical cytoplasmic protein required for transducing the Fz signal. At the interface between pre-R3 and pre-R4, Fz/Dsh is at the pre-R3 plasma membrane, where Dsh may directly inhibit Notch receptor activation in R3. The asymmetrically localized Fz/Dsh complex may also amplify the difference in Fz activation between the two cells through a feedback loop.
Here, we discover a unique Ral-dependent pathway by which Fz/PCP signaling leads to asymmetric Notch activation in R4. We show that in direct response to Fz activation, Ral expression in pre-R3 represses Notch activation, thereby biasing pre-R3 to become the Delta signaling cell. Moreover, we found unexpectedly that Ral prevents Notch activation that occurs independent of ligand binding. Thus, Ral regulates Notch signaling, and ligand-independent Notch activation is a target of regulation during cell patterning.
MATERIALS AND METHODS
Drosophila strains
The following alleles were used in this work. FlyBase id numbers, when available, are in parentheses. Chromosomes used are indicated in the figure legends. RalEE1 (FBal0197295); RalPG69 (FBal0130802); RalPG89 (FBal0130801); fzP21 (FBal0004937); fz2C1 (FBal0102708); fafFO8 (FBal0031258); neur1 (FBal0012940); Dl9P (FBal0002474); N5419 (FBab0000564); lqfFDD9 (FBal0104483); lqfAG (FBal0104486); Act5C-gal4 (FBti0012293); ey-gal4 (FBti0012711); ro-gal4 (E. Overstreet, PhD thesis, University of Texas at Austin, 2005); GMR-gal4 (FBti0072862); Act>stop>gal4 (from N.-S. Moon, McGill University, Quebec, Canada); tub-gal4 (from G. Struhl, Columbia University, New York, USA); tub-gal80 (FBti0012693, FBti0012683), UAS-flp (FBti0012285); UAS-Ralwt (FBal0101574); UAS-RalCA (FBal0101576); UAS-RalRNAi (VDRC# 105296); UAS-ngfp (on X, FBti0012492, FBti0012493); UAS-nlacZ (FBtp0001611); ubi-ngfp (FBti0015575, FBti0016102); sev-fz (FBal0082914); m∂-lacZ (FBtp0010977); ro-gfp (Overstreet et al., 2004); hs-N+-GV3 (FBal0090683); hs-NSev11-GV3 (Struhl and Adachi, 1998); FRT19A (FBti0000870); FRT82B (FBti0002074); FRT2A (FBti0002046); eyFLP (FBti0015984, FBti0015982); eyFLP2 (from B. Dickson, IMP, Vienna, Austria); hs-FLP (FBti0002044); and FM7, gfp (FBst0005193).
ro-gal4
gal4 DNA sequences were amplified from GMR-gal4 flies using primers that inserted AscI sites upstream of the start codon and downstream of the stop codon: 5′-ggcgcgccATGAAGCTACTGTCTTCTATCG-3′ and 5′-ggcgcgccTTACTCTTTTTTTGGGTTTGG-3′. The 2.7 kb amplification product was ligated into pGEM (Stratagene, Santa Clara, CA, USA), its sequence verified, and the AscI restriction fragment was ligated into the AscI site of pRO (Huang and Fischer-Vize, 1996) (E. Overstreet, PhD thesis, University of Texas at Austin, 2005). The resulting plasmid was used to transform w1118 flies.
Imaging of eyes and larvae
Immunohistochemistry of third instar larval eye discs was performed as follows. Discs were fixed in PEMS buffer with 1.0% NP-40 for 15 minutes. Antibody treatment was as described previously (Lim et al., 2007) with modifications. Fixed eye discs were blocked for 2 hours at 4°C in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40 and 5 mg/ml BSA, and then incubated in primary antibody diluted in blocking solution overnight at 4°C. Discs were washed in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl and 0.5% NP-40 three times for 5 minutes, and incubated with secondary antibodies in washing solution for 2 hours at room temperature, and then washed three times for 5 minutes. Phalloidin was used as described previously (Chen et al., 2002). Eye discs were mounted in Vectashield (Vector, Burlingame, CA, USA) and viewed with a Leica TCSSP2 confocal microscope. Primary antibodies were: rat monoclonal anti-Elav supernatant at 1:2 [Developmental Studies Hybridoma Bank (DSHB)], mouse monoclonal anti-β-galactosidase at 1:50 from the DSHB, rabbit polyclonal anti-RalB (Proteintech Group, Chicago, IL, USA) at 1:100, rabbit anti-Svp at 1:100. Secondary antibodies (Molecular Probes, Carlsbad, CA, USA) used at 1:200 were: Alexa568-anti-rabbit, Alexa568-anti-mouse, Alexa488-anti-rabbit, Alexa633-anti-rat, Cy5-anti-rabbit. Plastic sectioning of adult eyes was as described (Tomlinson and Ready, 1987). Eye sections were photographed with a Zeiss Axioplan and Axiocam HRc. Adult external eyes were photographed with an Olympus SZX12 microscope equipped with a SPOT idea (Diagnostic Instruments, Sterling Heights, MI, USA) digital camera. Larvae expressing GFP were viewed with a Leica M165FC microscope and photographed with a Leica DFC420C camera. Images were processed with Adobe Photoshop CS3. Statistical analysis of eye sections was using the unpaired t-test, Prism 3.0 software.
GFP protein blot
For quantitation of GFP (see Fig. S3 in the supplementary material), animals were grown at 29°C, heat shocked as third instar larvae for 1 hour at 37°C, and allowed to recover for 2 hours at 29°C. To generate protein extracts, five males were homogenized in 100 μl ice-cold PBS and 100 μl of 2× loading buffer was added. The extract was boiled for 10 minutes and then microfuged for 10 minutes. Supernatants (13 μl) were subjected to 10% SDS-PAGE and western blotted. Primary antibodies were rabbit anti-GFP (ABchem) at 1:5000 and mAbE7 (DSHB) at 1:1000. Secondary antibodies were HRP-anti-mouse and HRP-anti-rabbit (Santa Cruz Biochem) at 1:2000. Signals were quantified with NIH Image J software.
RESULTS
Ral is required for R-cell specification and PCP in the eye
Three Ral alleles were used in this work: RalEE1, RalPG69 and RalPG89. RalEE1 is a mis-sense mutation that alters a nucleotide-binding site (Ser154→Leu154) (Eun et al., 2007). RalPG69 and RalPG89 are P-element insertions (gal4-expressing enhancer traps) in the 5′-UTR and the first intron of Ral, respectively (Ghiglione et al., 2008). Ral protein expression levels from the P alleles are reduced relative to wild type.
RalEE1/Y males or RalEE1 homozygous females are viable with morphological abnormalities, including reduced rough eyes, curved wings, and missing hairs and bristles (Eun et al., 2007). RalEE1 behaves like a hypomorphic allele. All aspects of the RalEE1 mutant phenotype in hemizygous males are complemented by Act5C-gal4;UAS-Ralwt (Eun et al., 2007). In addition, heterozygotes for RalEE1 and either of the lethal hypomorphic alleles RalPG69 or RalPG89 have a mutant phenotype similar to RalEE1 hemizygotes or homozygotes, that is complemented by Ral PG69; UAS-Ralwt (data not shown). Moreover, flies that express RalRNAi in the eye have defects similar to those in RalEE1 flies, and the defects are rescued to wild-type by overexpression of wild-type Ral (see Fig. S1 in the supplementary material).
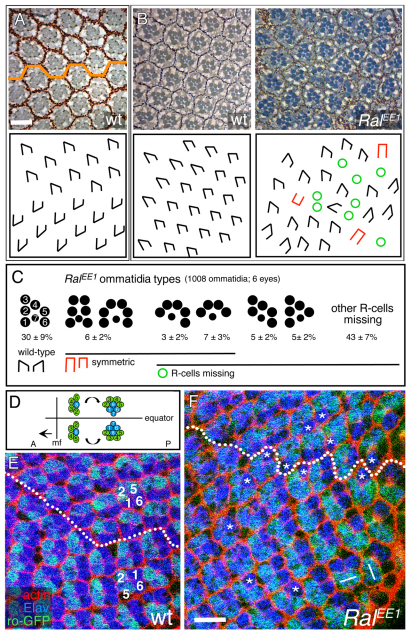
We analyzed the eyes of RalEE1/Y flies in detail. Wild-type adult eyes have ~800 facets, or ommatidia, each with eight photoreceptors (R cells) arranged in a trapezoid that is asymmetrical owing to the positions of R3 and R4 (Fig. 1A,D). In wild-type eyes, the trapezoids are perfectly aligned with one another. There are two chiral forms of ommatidia, mirror-image symmetrical through the equator that divides the eye into dorsal and ventral halves (Fig. 1A). Adult ommatidia of RalEE1/Y eyes had a variety of defects, including loss of R cells, loss of R3/R4 asymmetry and defects in orientation with respect to the equator (Fig. 1B,C). Adult eyes of RalEE1/RalPG69 and RalEE1/RalPG89 were similar to RalEE1/Y (data not shown). We examined RalEE1/Y eye discs to determine whether or not the adult eye abnormalities were due to defects in early development. The eye disc is a monolayer epithelium in which ommatidia assemble stepwise posterior to the morphogenetic furrow as it travels across the disc from posterior to anterior (Wolff and Ready, 1993). Five R-cell precursors (R8, R2/5, R3/4) emerge as a pre-cluster, and then R1/6 and R7 are recruited from the remaining pool of undifferentiated cells (Fig. 1D). Assembling ommatidia normally rotate in mirror-image reflection with respect to the equator (Fig. 1D). In RalEE1/Y eye discs, many ommatidia rotate either too much or too little, and R1 or R6 are frequently absent (Fig. 1E,F). Similar observations were made with RalEE1/RalPG69 and RalEE1/RalPG89 eye discs (data not shown). We conclude that Ral is required for patterning early in eye disc development.
Fig. 1.
Ral mutant eye phenotype. (A) An apical tangential section through a wild-type adult eye is shown at the top. The orange line marks the equator. The diagram beneath indicates the facet orientations. (B) (Top) Sections of wild-type and RalEE1/Y eyes. The diagrams beneath indicate facet orientations and mutant phenotypes. Symbols are defined in C. (C) Quantification of classes of RalEE1/Y ommatidia. Reverse orientation facets were not scored because in the context of all of the aberrant ommatidia, it was often difficult to locate the equator. (D) Diagram of five R-cell pre-clusters in a third instar larval eye disc and rotation with respect to the equator. A, anterior; P, posterior; mf, morphogenetic furrow, moving in the direction of arrow. (E,F) Third instar larval eye discs expressing GFP in R2/5 and R3/4, and immunolabeled with anti-Elav (R-cell nuclei) and phalloidin (actin). The genotypes are ro-gfp (wild-type) and RalEE1/Y; ro-gfp. The dotted line is the equator, and the morphogenetic furrow is leftward. Numbers indicate R2/R5 and R1/R6. Asterisks are ommatidia in which one of the R1/R6 pair are absent. The lines indicate misrotated ommatidia. Scale bar: in A, 20 μm for A,B; in F, 10 μm for E,F.
Ral eye defects and genetic interactions suggest that Ral regulates Notch signaling
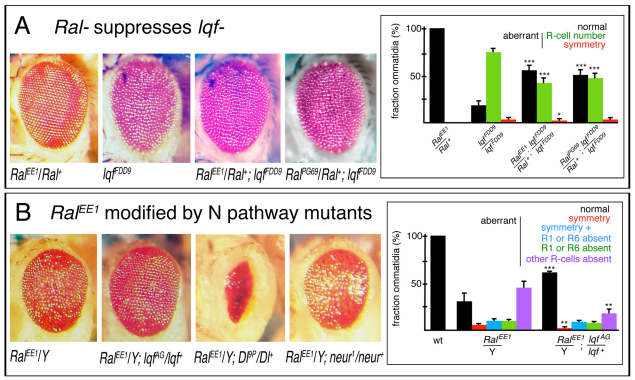
RalEE1 was identified in a mutagenesis screen for dominant enhancers of the rough eye caused by epsin overexpression (Eun et al., 2007). Epsin is an endocytic protein required in Notch signaling cells for ligand endocytosis and signaling (Overstreet et al., 2004; Wang and Struhl, 2004; Wang and Struhl, 2005). Loss-of-function alleles of genes acting in Notch signaling both positively and negatively were identified in the screen (Eun et al., 2007). To determine whether Ral plays a role in Notch signaling normally, we tested for genetic interactions between Ral and lqf loss-of-function mutations. Ral− mutations were dominant suppressors of lqf hypomorphic eye phenotypes (Fig. 2A), and the RalEE1 eye phenotype was suppressed by lqf (Fig. 2B). These results imply that Ral regulates Notch signaling, and further suggest that Ral activity opposes Notch activation.
Fig. 2.
Genetic interactions between Ral and Notch pathway mutants. (A) External adult eyes of the genotypes indicated are shown. Ral− heterozygous eyes are like wild type (smooth, left), lqfFDD9 eyes are rough and although this is not obvious in the photographs, eyes with both mutations are more like wild type than like lqfFDD9. The graph shows quantitative analysis of the wild-type and mutant ommatidia observed in tangential adult eye sections. For each genotype, the data were obtained from five eyes and 600-900 ommatidia. The symmetric ommatidia were scorable only in ommatidia with normal numbers of R cells. ***P<0.0001, *P<0.02; unpaired t-test. Data are mean + s.e.m. (B) External adult eyes of the indicated genotypes are shown. Ral− hemizygous eyes (left) are rough. They are made smoother by heterozygous loss-of-function of lqf, and rougher by heterozygous loss-of-function of Dl or neur. The suppressive effect of lqf is quantified on the right. RalEE1/Y data are from Fig. 1 and RalEE1/Y; lqfAG/lqf+ data were obtained from tangential sections of 732 ommatidia in four eyes. ***P<0.0005, **P<0.003; unpaired t-test. Data are mean + s.e.m.
Two additional observations supported the idea that Ral is a negative regulator of Notch signaling. First, N5419 (a null allele) was a dominant enhancer of the eye defects caused by expression of constitutively active Ral (Sawamoto et al., 1999) during eye development (ey-gal4, GMR-gal4; UAS-RalCA) (see Fig. S2 in the supplementary material). Second, the RalEE1 ommatidial defects described above are similar to those observed when there is too much Notch activity. Normal R3/R4 asymmetry (and rotation) results from Notch activation in the R4 precursor by Delta in the R3 precursor (Fanto and Mlodzik, 1999; Cooper and Bray, 1999; Tomlinson and Struhl, 1999). Notch activation in both R3/4 precursors results in equivalent symmetric R3/4 cells (Fanto and Mlodzik, 1999; Cooper and Bray, 1999), and overactive Notch in R1/6 precursors results in their failure to differentiate as R cells (Cooper and Bray, 1999). Too little Notch activation early in eye development likewise results in symmetric R3/4 cells (Fanto and Mlodzik, 1999; Cooper and Bray, 1999) and also extra R3/4 cells due to the failure of Notch activation in surplus precluster cells (Cagan and Ready, 1989; Overstreet et al., 2004). Extra R3/4 cells are not observed in RalEE1 ommatidia (Fig. 1B), suggesting that in Ral mutants, the Notch pathway is generally overactive.
In contrast to the suppressive interactions between RalEE1 and lqf−, we were surprised to find that the RalEE1 eye roughness was dominantly enhanced by loss-of-function mutations in two other Notch pathway genes, Dl and neur (Fig. 2B). These results suggest that Ral promotes Notch signaling. Taken together, the genetic interactions led us to conclude that Ral regulates Notch signaling, but in a complex manner. Further experiments described below that illuminated the role of Ral in Notch signaling also suggested resolutions to this paradox, and this will be explained below.
Ral prevents ligand-independent Notch activation
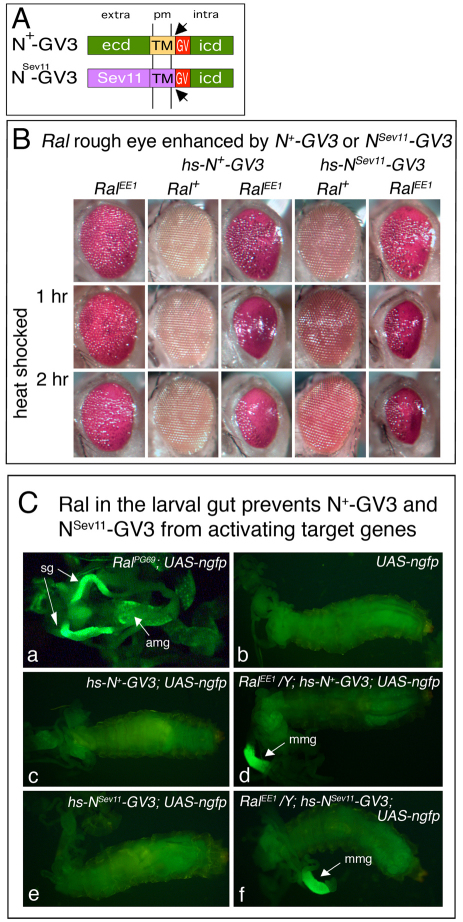
To examine further the idea that Ral represses Notch activation, we tested for genetic interactions between RalEE1 and transgenes that overexpress two different forms of the Notch receptor, called N+-GV3 and NSev11-GV3, under heat-shock control (Struhl and Adachi, 1998) (Fig. 3A). When Notch is activated, two proteolytic cleavages occur; an ADAM metalloprotease cleaves the extracellular domain, and then Presenilin cleaves within the transmembrane domain to generate a cytoplasmic fragment called Nicd (icd, intracellular domain) that travels to the nucleus and derepresses target gene transcription (Bray, 2006). N+-GV3 contains Gal4/VP16 within its Nicd fragment (Nicd-GV3). N+-GV3 functions in the same way as wild-type Notch in that the transgene complements N− mutations, and also Nicd-GV3 activates transcription of UAS-lacZ in response to ligand binding (Struhl and Adachi, 1998). NSev11-GV3 is an altered version of N+-GV3, in which the Notch extracellular and transmembrane domains were replaced by a truncated version of those domains of the Sevenless receptor. NSev11-GV3 thus cannot bind Notch ligands, and therefore it does not normally activate UAS-lacZ (Struhl and Adachi, 1998). We used both forms of the Notch receptor in order to determine whether the interaction between Ral and Notch required the Notch extracellular domain and, thus, ligand binding.
Fig. 3.
Interactions between Ral, N+-GV3 and NSev11-GV3. (A) Diagrams of the protein products of the hs-N+-GV3 and hs-NSev11-GV3 transgenes (Struhl and Adachi 1998). ecd, extracellular domain; icd, intracellular domain; TM, transmembrane domain; pm, plasma membrane; GV, Gal4/VP16; Sev11, truncated extracellular and TM domains of Sevenless receptor; arrow, cleavage site that generates Nicd-GV3. (B) External eyes of the genotypes indicated at the top are shown. The flies were either not heat-shocked, or else heat-shocked as third instar larvae for 1 or 2 hours at 37oC to express the hs-N+-GV3 or hs-NSev11-GV3 transgene. Neither N+-GV3 nor NSev11-GV3 causes eye roughness, but each enhances the roughness of RalEE1/Y eyes. (C) Dissected third instar larvae with gut extruded from cuticle, photographed to visualize GFP fluorescence. Ral-driven GFP expression (part a) is visible in the salivary glands (sg) and anterior midgut (amg), and mainly further posterior in the middle midgut (mmg) in parts d and f. GFP expression was quantified on protein blots (see Fig. S3 in the supplementary material).
If Ral represses Notch activation, then N+-GV3 overexpression would be expected to enhance the RalEE1 rough eye, and we found that it does (Fig. 3B). Remarkably, we also found that NSev11-GV3 overexpression had a similar effect (Fig. 3B). These results suggest that in RalEE1 cells, both N+-GV3 and NSev11-GV3 receptors are activated. To test this, we assayed the expression of UAS-ngfp in Ral+ or RalEE1 larvae that express either N+-GV3 or NSev11-GV3. We observed little or no GFP in Ral+ larvae (Fig. 3C, parts c,e), but in Ral− larvae, there were high levels of GFP in the midgut (Fig. 3C, parts d,f) (see also Fig. S3 in the supplementary material). Ral expression is reportedly elevated in the larval gut (Tweedie et al., 2009), and Ral is required for intestinal antibacterial immunity (Cronin et al., 2009). Consistent with these reports, we observed high levels of GFP in the midguts of RalPG69; UAS-ngfp larvae (Fig. 3C, part a). We conclude that Ral blocks ligand-independent activation of Notch.
Ral activity in R3 promotes R3/R4 asymmetry
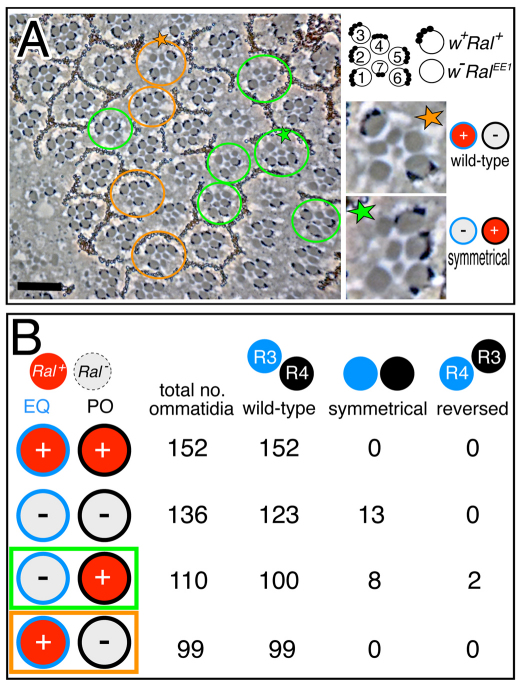
To understand the developmental context in which Ral activity is important for eye patterning, we focused on R3/R4 asymmetry, which is affected in Ral− mutants (Fig. 1B,C). Notch activation in the polar cell (pre-R4) by Delta in the equatorial cell (pre-R3) distinguishes R3 and R4 (Fanto and Mlodzik, 1999; Cooper and Bray, 1999; Tomlinson and Struhl, 1999). As Ral activity antagonizes Notch activation, the simple expectation is that Ral is required in the signaling cell, R3. To determine whether there is a requirement for Ral in either R3 or R4, we used FLP/FRT-induced mitotic recombination to induce white−-marked (w−) RalEE1 homozygous clones in RalEE1/Ral+ eyes, thereby generating ommatidia mosaic for w− RalEE1 and w+ Ral+ cells at the clone borders (Fig. 4A). We scored the phenotypes of ommatidia at the clone borders in which one of the R3/R4 pair was Ral+ and one was RalEE1, and also of ommatidia in which both cells of the R3/4 pair were either Ral+ or RalEE1 (Fig. 4B). When both of the R3/R4 pair were Ral+, all of the facets were asymmetrical and had normal chirality; the equatorial cell always became R3 and the polar cell always became R4. When both were RalEE1, ~10% of ommatidia were symmetrical, meaning that R3 and R4 were not distinguished. In ommatidia where one of the R3/R4 pair was Ral+ and the other RalEE1, different results were observed depending on whether the equatorial cell or the polar cell was Ral+. When the equatorial cell was Ral+ (and the polar cell RalEE1), all of the ommatidia were wild type. By contrast, when the polar cell was Ral+ (and the equatorial cell RalEE1), ~7% were symmetrical and ~2% had reversed chirality, meaning that the polar cell became R3 and the equatorial cell became R4. These results lead to two conclusions. First and most significantly, Ral+ functions in the equatorial cell (pre-R3) to promote asymmetric R3/R4 differentiation. We know this because whenever the equatorial cell was Ral+, R3 and R4 were always properly asymmetrical, but when the equatorial cell was Ral−, symmetrical R3/R4 cells were sometimes observed. Second, having higher Ral+ activity (at least the difference between RalEE1 and Ral+) is insufficient to ensure that an R3/R4 precursor will always become R3. We know this because in ommatidia where the polar cell was Ral+ and the equatorial cell was RalEE1, polar cells did occasionally reverse their normal differentiation and become R3, but more often the R3/R4 pair was either symmetrical or wild type.
Fig. 4.
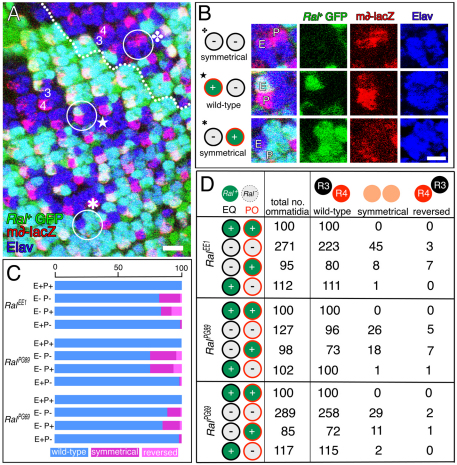
Analysis of adult ommatidia where pre-R3/pre-R4 are mosaic for Ral+ or Ral−. (A) An apical tangential section through a w− Ral−/w− Ral− clone in a w− Ral− / w+ Ral+ eye. The genotype is: w RalEE1 FRT19A/FRT19A; eyFLP/+. Circled ommatidia have mosaic R3/R4 cells; orange are EQ+/PO− (all asymmetrical) and green are EQ−/PO+ (the starred one is symmetrical). Scale bar: 20 μm. (B) Pooled results from 13 different eye clones are shown. EQ, equatorial cell (pre-R3); PO, polar cell (pre-R4).
Ral activity in R3 promotes asymmetric Notch activation in R4
We have shown above that Ral+ activity in R3 influences R3/R4 asymmetry, but does it do so through an effect on Delta/Notch signaling? Notch activation in the R3/R4 pair may be monitored by the expression of a transgene called m∂-lacZ, in which the transcriptional control sequences of the Notch target gene E(spl)m∂ drives expression of lacZ (Cooper and Bray, 1999). In wild-type third instar larval eye discs, m∂-lacZ is expressed mainly in the polar cell, which becomes R4 (Cooper and Bray, 1999) (Fig. 5A). To determine whether Ral+ activity in either the equatorial (pre-R3) or polar cell (pre-R4) affects Notch activation (m∂-lacZ expression), we generated developing ommatidia mosaic for gfp+ Ral+ and gfp− Ral− cells by inducing gfp− Ral− clones (RalEE1, RalPG69 or RalPG89) in gfp+ Ral+/Ral− eye discs. We scored m∂-lacZ expression in mosaic facets in which the polar and equatorial cells were both Ral+, both Ral−, or where one of the R3/R4 pair was Ral− and one was Ral+ (Fig. 5B-D). We found that when both the equatorial and polar cells were Ral+, m∂-lacZ was expressed in R4. By contrast, when both cells of the R3/R4 pair were Ral−, m∂-lacZ expression was often symmetrical (absent or at low levels in both cells), or occasionally the pattern of m∂-lacZ expression was reversed, meaning that the equatorial cell expressed m∂-lacZ and the polar cell did not. These results indicate that Ral+ activity in the R3/R4 pair does affect Notch activation. In addition, the pattern of m∂-lacZ expression was almost always wild-type when the equatorial cell was Ral+ and the polar cell Ral−. By contrast, when the equatorial cell was Ral− and the polar cell Ral+, m∂-lacZ expression was often symmetrical or reversed. We conclude that Ral+ activity in the equatorial cell (pre-R3) promotes asymmetric Notch activation in the polar cell (pre-R4).
Fig. 5.
Analysis of developing larval ommatidia where pre-R3/pre-R4 are mosaic for Ral+ or Ral−. (A) A RalEE1/gfp+ eye disc with RalEE1 homozygous (gfp−) clones is shown. The disc expresses m∂-lacZ and is immunolabeled with anti-β-gal (Notch activation) and anti-Elav (R-cell nuclei). The genotype is: RalEE1 FRT19A/ubi-gfp FRT19A; ey-gal4, UAS-flp/+; m∂-lacZ/+. The broken line is the equator, the furrow is on the left. The numbers are R3/R4 cells. Circled ommatidia are enlarged in B. Scale bar: 10 μm. (B) Enlargements of a Ral−/Ral− (E/P) ommatidium, and two kinds of mosaics are shown. E, equatorial cell; P, polar cell. Scale bar: 5 μm. (C) Fractions of normally constructed (wild-type) and mutant (symmetrical or reversed) ommatidia in each of the four genotypic classes indicated. The scale at the top is in percent. E, equatorial cell; P, polar cell; + is Ral+; − is Ral−. (D) Raw data for graph in C. EQ, equatorial cell; PO, polar cell.
The role of Ral in R3 clarifies how different loss-of-function mutations in genes that promote Delta signaling, Delta and neur versus lqf, can have opposite effects on the Ral mutant phenotype. Ral, Delta and neur are all required in the pre-R3 cell, where they bias pre-R3 to become the Delta signaler. Pre-R3 is sensitive to the levels of activity of all three genes, and so the observation that Delta or neur mutations enhance Ral mutations makes sense in this context. Why does lqf interact with Ral in the opposite way? One possibility is that pre-R4 is more sensitive to Lqf levels than pre-R3 is, and so the major effect of lqf mutation is not in pre-R3, but in pre-R4. Unlike Delta and neur, lqf is not upregulated in pre-R3 (B.C. and J.A.F., unpublished observations). Perhaps the lower levels of Delta and Neur in pre-R4 render pre-R4 more sensitive than pre-R3 to the levels of Lqf. If so, the negative effect of lowering the lqf gene dose on the ability of a cell to become the signaler would be more significant in pre-R4 than in pre-R3. In this scenario, Ral− or lqf− mutations would have opposite effects on R3/R4 asymmetry, and would be expected to suppress each other. Alternatively, Lqf might antagonize Ral activity directly in R3 by promoting ligand-independent Notch activation. The latter possibility may be tested with additional experiments. If the role of Ral in other cell fate decisions in the eye is similar to its role in R3/R4, then this kind of logic could explain the effects on overall eye roughness observed in various mutant combinations.
Ral expression is enriched in R3 and depends on Frizzled near the morphogenetic furrow
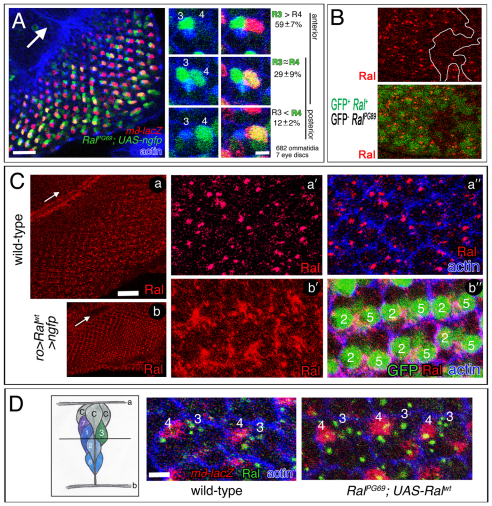
The results above indicate a requirement for Ral+ in the equatorial cell (pre-R3). We were curious to know whether Ral expression is ubiquitous or spatially restricted. To monitor Ral transcriptional activation, we used RalPG69, the gal4-expressing enhancer trap in the 5′-UTR of Ral, driving expression of UAS-ngfp. We found that GFP was expressed in all R-cells, but in the majority of ommatidia, GFP was enriched in R3 beginning at approximately row 4 posterior to the furrow (Fig. 6A). In ~12% of ommatidia, GFP levels were higher in R4 than in R3 (Fig. 6A), but all of these ommatidia were posterior to row 7. As m∂-lacZ expression in R4, which is indicative of R3/R4 specification, normally begins at row 3 or 4 (Fanto and Mlodzik, 1999; Cooper and Bray, 1999), RalPG69 activity is generally elevated in pre-R3 at the time when R3 and R4 are specified. (We expect a delay of about one row in RalPG69 due to the Gal4 intermediate.)
Fig. 6.
Ral expression in eye discs. (A) An eye disc is shown on the left that expresses m∂-lacZ and ngfp under the control of a Ral enhancer trap, and immunolabeled with anti-β-gal and phalloidin. The genotype is: RalPG69/+; m∂-lacZ/UAS-ngfp. Enlargements of the three different classes of ommatidia indicated are on the right. Scale bars: in the large panel and in B, 20 μm; in the small panels, 5 μm. Arrow indicates morphogenetic furrow. (B) A RalPG89/Ral+ eye disc containing a clone of RalPG89/RalPG89 cells (outlined at top), immunolabeled with anti-Ral. The genotype is RalPG89 FRT19A/ubi-gfp FRT19A; ey-gal4, UAS-flp/+. (C) Wild-type eye discs and eye discs that overexpress Ral and ngfp in R2/R5 and R3/R4 under the control of ro-gal4 are shown. The genotype is: ro-gal4/+; UAS-Ral/UAS-ngfp. The discs are immunolabeled with anti-Ral and phalloidin. Scale bar: 50 μm in a,b; 5 μm in a′,a″,b′,b″. Arrows indicate morphogenetic furrow. (D) A z-section of a developing ommatidium. A, apical membrane; b, basal membrane; c, cone cell; numbers are R-cells. The horizontal line represents the depth of the xy images on the right. A Ral+ (wild-type) eye disc that expresses m∂-lacZ and an eye disc that also overexpresses Ral under control of a Ral enhancer trap (RalPG69; UAS-Ral) are shown. The genotypes are: m∂-lacZ/+ (wild type) and RalPG69/+; UAS-Ral/m∂-lacZ. Each is immunolabeled with anti-β-gal, anti-Ral and phalloidin. The numbers are R3 and R4. We counted the number of R3/R4 pairs in which there were more Ral+ puncta in R3 (R3>R4), where the numbers were similar (R3~R4), and where there were more in R4 (R4>R3) in wild-type and RalPG69; UAS-Ral eye discs. In five wild-type discs: R3>R4 (211/290), R3~R4 (31/290), R4>R3 (48/290). In six RalPG69; UAS-Ral discs: R3>R4 (161/219), R3~R4 (21/219), R4>R3 (37/219). Scale bar: 5 μm.
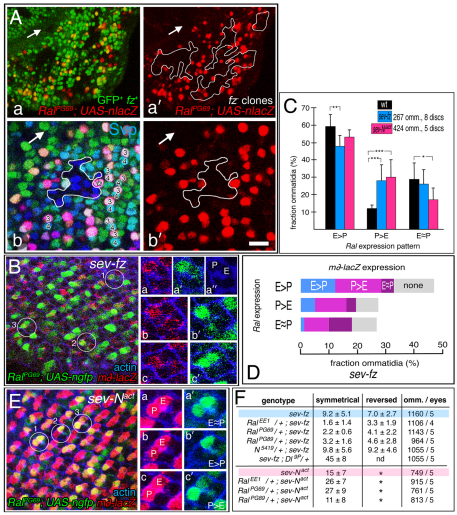
Specification of the equatorial cell as R3 depends on Fz activation (Zheng et al., 1995; Tomlinson and Struhl, 1999; Fanto and Mlodzik, 1999; Cooper and Bray, 1999). In fz− eyes, ommatidia are symmetrical, or their chirality is randomized (wild-type or reversed). Fz activation increases transcription of Delta and neur (Fanto and Mlodzik, 1999; del Alamo and Mlodzik, 2006) and may also repress Notch receptor activation in the equatorial cell (Strutt et al., 2002). To determine whether RalPG69 activity depends on Fz signaling, we monitored β-galactosidase (β-gal) in fz− clones with RalPG69; UAS-nlacZ. β-Gal expression was reduced in fz− clones, most severely near the morphogenetic furrow, where R3 and R4 are first distinguished (Fig. 7A, parts a-b′). We also observed non-autonomous repressive effects of fz− clones on Ral expression outside of the clones (Fig. 7A, parts a-b′). Moreover, at the borders of clones, when one cell of a mosaic (fz+/fz−) R3/R4 pair expressed β-galactosidase, it was usually the fz+ cell (15/21 pairs in eight clones) (Fig. 7A, parts b,b′), and the six exceptions were at the posterior of the eye disc.
Fig. 7.
Control of Ral expression by Fz and Notch. (A) Eye discs are shown containing fz− gfp− clones (white outlines). The discs express nuclear β-gal under Ral control and were immunolabeled with anti-β-gal and anti-Svp (labels R3 and R4). The genotype is: RalPG69/eyFLP; UAS-nlacZ/+; fzP21 fz2C1 FRT2A/ubi-gfp FRT2A. Arrows indicate the furrow. The disc in a,a′ contains several clones. In b,b′, a single clone from a different disc is shown enlarged. R3 and R4 are indicated. The asterisk indicates a fz+ R4 (β-gal+) of a mosaic pair where the R3 is fz− (β-gal−). (B) An eye disc that expresses sev-fz, m∂-lacZ and ngfp under Ral control, immunolabeled with anti-β-gal and phalloidin, is shown. The genotype is: RalPG69/+; sev-fz/+; m∂-lacZ/UAS-ngfp. Enlargements of the three circled ommatidia are on the right. The furrow is on the right. E, equatorial cell; P, polar cell. In a and b, Ral expression is P>E, and in c, it is E~P. (C) Quantification of the three types of Ral expression patterns in ommatidia in sev-fz or sev-Nact discs. ***P<0.001, **P<0.01, *P<0.04. Data are mean + s.e.m. (D) Quantification of the four types of m∂-lacZ expression pattern within each of the three types of Ral expression pattern in the sev-fz discs in C. (E) An eye disc is shown that expresses sev-Nact, m∂-lacZ and ngfp under Ral control, immunolabeled with anti-β-gal and phalloidin. Circled ommatidia representing each of the three classes of Ral expression pattern are enlarged on the right. The furrow is on the left. The genotype is: RalPG69/+; sev-Nact/+; m∂-lacZ/UAS-ngfp. (F) A table showing quantification of dominant genetic interactions between Ral mutants, and also null alleles of Notch and Delta, with sev-fz, and Ral mutants with sev-Nact. Data were obtained from sections of adult eyes. omm., number of ommatidia; nd, not determined; *, reversed ommatidia could not be scored because the eye field was too disorganized. Scale bar: 40 μm in A, parts a,a′; 20 μm in A, parts b,b′, and in B,E; 10 μm in the enlargements in B,E.
As expression of UAS-Ralwt under RalPG69 control complements Ral− mutants, the activity of RalPG69 observed probably mirrors, at least in part, the normal Ral transcription pattern. To test this assumption, we examined Ral protein in eye discs using a polyclonal antibody to human RalB [Drosophila Ral and human RalB are identical in 148/201 amino acids, and a different antibody to human Ral B was used to recognize Drosophila Ral in ovaries and on protein blots (Balakireva et al., 2006; Ghiglione et al., 2008)]. In wild-type eye discs, the antibody labeled puncta posterior to the furrow (Fig. 6C, parts a,a′). Although a Ral protein null allele to use as a control is unavailable, several experiments lead us to conclude that the antibody recognizes Ral specifically in the eye. First, the antibody signal was strikingly lower in RalPG89 homozygous clones than in surrounding heterozygous tissue (Fig. 6B). Second, in eye discs that overexpress Ral in a subset of R cells (R2/5, R3/4) using ro-gal4; UAS-Ralwt, highly elevated signal was detected in R2/5 and R3/4 (Fig. 6C, parts b,b′). Third, the pattern of antibody labeling resembles closely the pattern of expression of GFP from RalPG69; UAS-ngfp. Ral antibody signal begins posterior to the furrow in approximately row 3 or 4 (Fig. 6C, parts a), and appears generally elevated in R3 (Fig. 6D). In wild-type eye discs, or in eye discs that overexpress Ral using RalPG69; UAS-Ralwt, Ral protein is in basal puncta (Fig. 6C, parts a′,a″), that in most ommatidia appear concentrated in R3 (Fig. 6D and legend).
The results above indicate that near the morphogenetic furrow, Ral expression is controlled by Fz. We tested this idea further by overexpressing fz in the R3/R4 pair using a sevenless expression vector construct, sev-fz. Expression of sev-fz results in R3/4 symmetry or random chirality, because the equalization of and/or excess of Fz activity in the R3/4 pair disrupts asymmetric Notch activation in the polar cell (pre-R4) (Fanto and Mlodzik, 1999; Cooper and Bray, 1999; Tomlinson and Struhl, 1999). We found that the normal pattern of RalPG69; UAS-ngfp expression was disrupted in eye discs expressing sev-fz (Fig. 7B,C). RalPG69; UAS-ngfp expression was enriched in the equatorial cell (pre-R3) less often than in wild-type discs, and enriched in the polar cell (pre-R4) more often than in wild type (Fig. 7C). We conclude that Ral transcriptional control is downstream of Fz.
Frizzled control of asymmetric Ral expression is not through Notch
Enrichment of Ral transcription in R3 versus R4 near the furrow could be a direct effect of Fz signaling, and a reflection of more Fz signaling in R3 than R4. Alternatively, asymmetric Ral expression could be downstream of Notch activation. If so, Ral enrichment in R3 could reflect that Notch activation in R4 represses Ral transcription directly in R4 and/or that Notch activation in R4 activates Ral transcription in R3 through a feedback mechanism. One observation suggests that Ral expression could be controlled by Notch; in sev-fz discs, m∂-lacZ (Notch activation) was generally depressed (Fig. 7B), and Ral had an increased tendency to be expressed in the polar cell (pre-R4) (Fig. 7D). However, we observed further that in sev-fz discs, m∂-lacZ and RalPG69; UAS-ngfp were often expressed in the same cell (Fig. 7D). Moreover, there was no tendency for cells that express m∂-lacZ not to express RalPG69; UAS-ngfp (Fig. 7D). We also generated discs in which the R3/R4 pair both express constitutively active Notch (sev-Nact), which renders them symmetrical (Fanto and Mlodzik, 1999; Cooper and Bray, 1999). Although the pattern of RalPG69; UAS-ngfp is disrupted somewhat, Ral expression is not depressed (Fig. 7E), and Ral has a greater tendency than in wild type to be expressed asymmetrically (Fig. 7C). Finally, we find that Ral− mutations suppress sev-fz defects, but fail to suppress (and in fact enhance) sev-Nact defects (Fig. 7F). This means that Fz in the R3/4 pair requires Ral even when Notch signaling is depressed and the R3/4 pair are symmetrical, but Notch does not. We conclude that Fz activation controls Ral transcription directly, not through Notch activation.
Ral-mediated Notch inhibition is one of several Fz-dependent pathways that control R3/R4 asymmetry
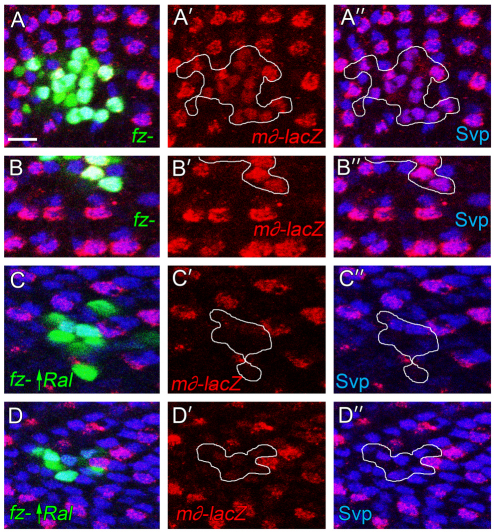
The effects on R3/R4 asymmetry of losing Ral in one or both cells are small; symmetry defects were observed in these experiments in 10-30% of the R3/R4 pairs (Fig. 1C; Fig. 4; Fig. 5). In contrast to the results with Ral (Fig. 5), in ommatidia where R3/R4 are mosaic for Delta, the Delta+ cell always becomes R3 (Tomlinson and Struhl, 1999). The small effects observed for Ral could result, at least in part, from the Ral+ function remaining in the three hypomorphic mutants we used. However, similar weak effects were observed in analogous experiments using a strong neur loss-of-function allele (del Alamo and Mlodzik, 2006). This suggests that RalEE1 may have an incompletely penetrant effect on R3/R4 asymmetry primarily because Ral works in only one or a subset of distinct Fz-dependent pathways that bias the pre-R3 cell to become the Delta signaler. Consistent with this interpretation, we find that overexpression of Ral (or RalCA, not shown) in both R3 and R4 in clones of otherwise wild-type eye disc cells had no effect on the pattern of Notch activation; m∂-lacZ is still expressed specifically in the polar cell (pre-R4) (Fig. 8A,A′). Even in mosaic R3/R4 pairs at the clone border in which RalCA is overexpressed in pre-R4 and not in pre-R3, m∂-lacZ was nearly always expressed in the polar cell (Fig. 8B). Similarly, Ral or RalCA overexpression in both R3 and R4 with ro-gal4; 2XUAS-Ral had only a very subtle effect on R3/4 asymmetry in the adult eye (see Table S1 in the supplementary material). If Ral functioned in the sole pathway or in all Fz-dependent pathways for R3 specification, we would expect reversal of the normal Ral expression pattern in mosaics to reverse R3/R4 polarity, and equalization of Ral expression in R3 and R4 to result in R3/R4 symmetry. These effects were observed but they were very subtle. Strikingly similar results were obtained in analogous experiments with neur (del Alamo and Mlodzik, 2006). Moreover, we find that in fz− cell clones, where all the pathways downstream of Fz that normally bias R3 to become the signaling cell are absent, Ral overexpression now does determine R3 cell fate. In clones of fz− cells in the eye disc, both cells of the R3/R4 pair express m∂-lacZ, at reduced levels compared with wild-type R4 (the cells are both R4 or some fate in between R3 and R4) (Fig. 9A-B′). Ral overexpression in fz− R3/R4 precursor pairs abolished m∂-lacZ expression (both cells are now R3) (Fig. 9C-D″). We conclude that Ral functions in one of several Fz-dependent pathways that control R3/R4 asymmetry.
Fig. 8.
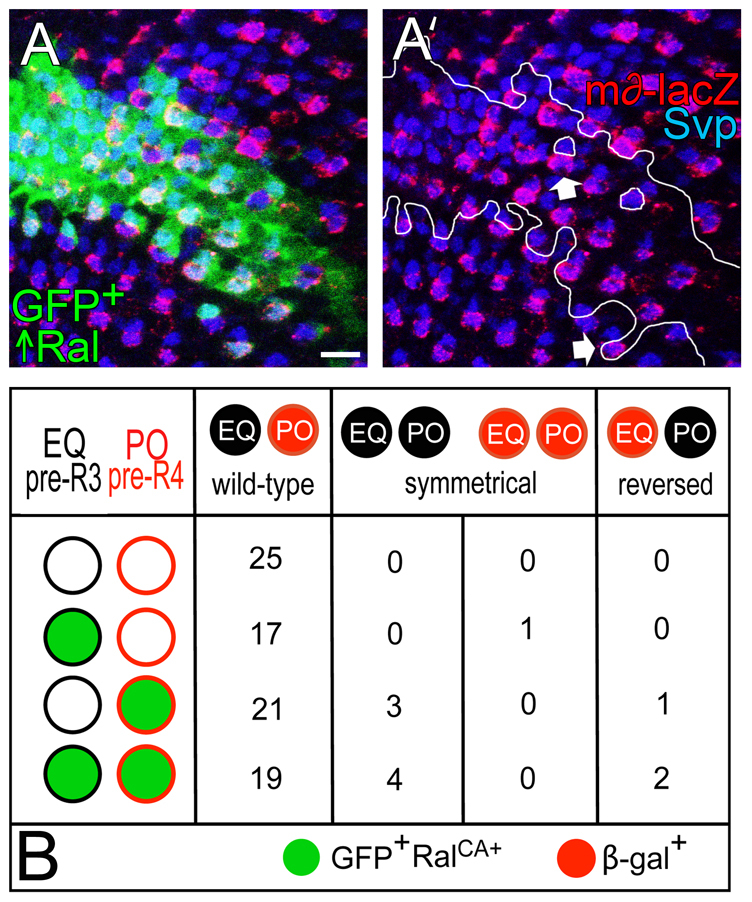
Ral overexpression in pre-R4 has only a subtle effect on R3/R4 determination. (A,A′) An eye disc is shown containing a gfp+ cell clone (outlined in A′) that overexpresses Ral. The eye disc also expresses m∂-lacZ and is immunolabeled with anti-Svp and anti-β-gal. The arrows indicate Ral-overexpressing (gfp+) R4s (β-gal+), the R3s of which (β-gal−) do not overexpress Ral (gfp−). The genotype is hs-flp, tub-gal4, UAS-gfp/+; UAS-Ralwt/m∂-lacZ; FRT82B/FRT82B tub-gal80. Larvae (2nd and 3rd instar) were heat-shocked for 1 hour at 37°C. (B) An analysis of R3/R4 determination in pairs mosaic for wild-type and RalCA-overexpressing cells. Pooled results from six mosaic eye discs are shown. EQ, equatorial cell; PO, polar cell. The genotype is hs-flp; Act5C>stoP>gal4, UAS-gfp/+; UAS-RalCA/m∂-lacZ. Scale bar: 10 μm.
Fig. 9.
Ral controls R3/R4 cell fate in fz− cells. (A-B″) Two separate gfp+ fz− clones (outlined) in an eye disc are shown in A-A″ and B-B″. The eye disc also expresses m∂-lacZ and is immunolabeled with anti-β-gal and anti-Svp. The genotype is hs-flp tub-gal4, UAS-gfp/+; m∂-lacZ/+; fzP21 fz2C1 FRT2A/tub-gal80 FRT2A. (C-D″) Two separate gfp+ fz− clones that overexpress Ral (outlined) in eye discs are shown in C-C″ and D-D″. The eye disc also expresses m∂-lacZ, and is immunolabeled with anti-β-gal and anti-Svp. The genotype is hs-flp tub-gal4, UAS-gfp/+; m∂-lacZ/UAS-Ralwt; fzP21 fz2C1 FRT2A/tub-gal80 FRT2A. Scale bar: 10 μm. To generate clones, larvae (2nd and 3rd instar) were heat-shocked for 1 hour at 37°C.
DISCUSSION
The results presented support the model for Ral function shown in Fig. 10. Ral transcription is upregulated in response to Fz activation. As Fz is activated more in the equatorial cell than the polar cell, Ral is enriched in the equatorial cell. Ral activity represses ligand-independent Notch activation, and thus biases the equatorial cell to become R3. One way in which ligand-independent Notch activation occurs is an accident when normal Notch trafficking is disrupted (Fortini and Bilder, 2009; Fortini, 2009). Notch receptor undergoes endocytosis and endosomal trafficking continually and mutations that block trafficking of late endosomes to the lysosome block Notch degradation and result in endosomal accumulation of Notch and ligand-independent activation (Fortini and Bilder, 2009; Fortini, 2009). One possibility is that the endosomal environment promotes production of Nicd by Presenilin cleavage. Ligand-independent Notch activation may also occur normally in the lysosomal membrane (Wilkin et al., 2008; Fortini and Bilder, 2009). Ral GTPase activity might block ligand-independent Notch activation by regulating Notch trafficking to the lysosome, or by inhibiting another process, such as endosomal acidification (Yan et al., 2009), Nicd production or nuclear translocation. The punctate appearance of Ral protein suggests the possibility that Ral may play a role in endosomal trafficking. Although further experiments are required to determine the precise mechanism of Ral function, we have shown that Ral, a protein that prevents ligand-independent Notch activation, is a target for regulation during pattern formation. Fz/PCP signaling upregulates Ral expression to ensure that ligand-independent Notch activation does not tip the scales in favor of pre-R3 becoming the signal receiving cell. Moreover, we have shown that prevention of ligand-independent Notch activation is not simply a constitutive process, but one that is modulated to ensure specific developmental outcomes.
Fig. 10.
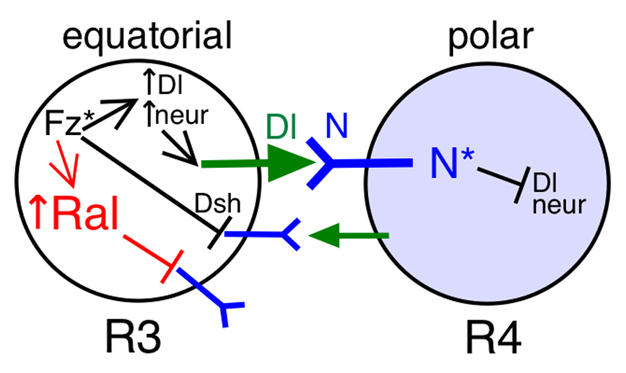
Model for Ral function in R3/R4 cell fate decision. Fz activation in the equatorial cell results in asymmetric Notch activation through two proposed mechanisms: (1) promotion of Delta signaling through transcriptional upregulation of Delta and neur, and their repression in the polar cell; and (2) direct repression of the Notch receptor through relocalization of a Fz/Dsh complex to the side of the equatorial cell plasma membrane adjacent to the polar cell. We have presented evidence for a distinct Ral-dependent mechanism in the equatorial cell. Ral transcription is upregulated in response to Fz activation, and Ral activity represses ligand-independent Notch activation. Notch activation in R4 does not repress Ral transcription in the polar cell.
Supplementary Material
Acknowledgements
We thank M. Balakireva, S. Bray, J. Camonis, B. Dickson, M. Fortini, H. Kramer, M. Mlodzik, N.-S. Moon, K. Sawamoto, G. Struhl, the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center for fly strains, and R. Cripps for anti-Svp. We are grateful to Paul Macdonald and Steve Vokes for use of their microscopes. This work was supported by a grant to J.A.F. from the US National Institutes for Child Health and Human Development (R01-HD30680). Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.056002/-/DC1
References
- Axelrod J. D. (2009). Progress and challenges in understanding planar cell polarity signaling. Semin. Cell Dev. Biol. 20, 964-971 [DOI] [PubMed] [Google Scholar]
- Balakireva M., Rosse C., Langevin J., Chien Y., Gho M., Gonzy-Treboul G., Voegeling-Lemaire S., Aresta S., Lepesant J.-A., Bellaiche Y., et al. (2006). The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 26, 8953-8963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signaling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678-689 [DOI] [PubMed] [Google Scholar]
- Cagan R. L., Ready D. F. (1989). Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 3, 1099-1112 [DOI] [PubMed] [Google Scholar]
- Camonis J. H., White M. A. (2005). Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol. 15, 327-332 [DOI] [PubMed] [Google Scholar]
- Cascone I., Rasim S., Ozdemir C., Del Nery E., Yeaman C., White M., Camonis J. (2008). Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 27, 2375-2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang B., Fischer J. A. (2002). A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of Fat facets. Genes Dev. 16, 289-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. W., Inoue M., Hsu S. C., Salltiel A. R. (2006). RalA-exocyst-dependent recycling endosome trafficking is required for the completion of cytokinesis. J. Biol. Chem. 281, 38609-38616 [DOI] [PubMed] [Google Scholar]
- Chien Y., Kim S., Bumeister R., Loo Y.-M., Kwon S. W., Johnson C. L., Balakireva M. G., Romeo Y., Kopelovich L., Gale M., Jr, et al. (2006). RalB GTPase-mediated activation of the IkB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127, 157-170 [DOI] [PubMed] [Google Scholar]
- Cooper M. T. D., Bray S. J. (1999). Frizzled regulation of Notch signaling polarizes cell fate in the Drosophila eye. Nature 397, 526-530 [DOI] [PubMed] [Google Scholar]
- Cronin S. J. F., Nehme N. T., Limmer S., Liegeois S., Pospisilik J. A., Schramek D., Leibbrandt A., de Matos Simoes R., Gruber S., Puc U., et al. (2009). Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325, 340-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Alamo D., Mlodzik M. (2006). Frizzled/PCP-dependent asymmetric neuralized expression determines R3/R4 cell fates in the Drosophila eye. Dev. Cell 11, 887-894 [DOI] [PubMed] [Google Scholar]
- Eun S. H., Lea K., Overstreet E., Stevens S., Lee J.-H., Fischer J. A. (2007). Identification of genes that interact with Drosophila liquid facets. Genetics 175, 1163-1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanto M., Mlodzik M. (1999). Asymmetric activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature 397, 523-526 [DOI] [PubMed] [Google Scholar]
- Feig L. A. (2003). Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 13, 419-425 [DOI] [PubMed] [Google Scholar]
- Fortini M. E. (2009). Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633-647 [DOI] [PubMed] [Google Scholar]
- Fortini M. E., Bilder D. (2009). Endocytic regulation of Notch signaling. Curr. Opin. Genet. Devel. 19, 323-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C., Devergne O., Cerezo D., Noselli S. (2008). Drosophila RalA is essential for the maintenance of Jak/Stat signaling in ovarian follicles. EMBO Rep. 9, 676-682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K., Kimura S., Takatsu H., Ohmae M., Kawano S., Kitamura H., Ito M., Watari H., Hazelett C. C., Yeaman C., et al. (2009). M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat. Cell Biol. 11, 1427-1432 [DOI] [PubMed] [Google Scholar]
- Huang Y., Fischer-Vize J. A. (1996). Undifferentiated cells in the developing Drosophila eye influence facet assembly and require the Fat facets ubiquitin-specific protease. Development 122, 3207-3216 [DOI] [PubMed] [Google Scholar]
- Klein T. J., Mlodzik M. (2005). Planar cell polarization: an emerging model points in the right direction. Annu. Rev. Cell Dev. Biol. 21, 155-176 [DOI] [PubMed] [Google Scholar]
- Lalli G. (2009). RalA and the exocyst complex influence neuronal polarity through PAR-3 and aPKC. J. Cell Sci. 122, 1499-1506 [DOI] [PubMed] [Google Scholar]
- Lawrence P. A., Struhl G., Casal J. (2007). Planar cell polarity: one or two pathways? Nat. Rev. Genet. 8, 555-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Lee O. K., Hsu Y. C., Singh A., Choi K. W. (2007). Drosophila TRAP230/240 are essential coactivators for Atonal in retinal neurogenesis. Dev. Biol. 308, 322-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T., Tanaka M., Nishina H., Murakami H., Yasunori K. (2008). RalA functions as an indispensable signal mediator for the nutrient-sensing system. J. Biol. Chem. 283, 35053-35059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko S., Henry D. O., Rosse C., Mirey G., Camonis J. H., White M. A. (2001). The exocyst is a Ral effector complex. Nat. Cell Biol. 4, 66-72 [DOI] [PubMed] [Google Scholar]
- Overstreet E., Fitch E., Fischer J. A. (2004). Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development 131, 5355-5366 [DOI] [PubMed] [Google Scholar]
- Sawamoto K., Yamada C., Kishida S., Hirota Y., Taguchi A., Kikuchi A., Okana H. (1999). Ectopic expression of constitutively activated Ral GTPase inhibits cell shape changes during Drosophila eye development. Oncogene 18, 1967-1974 [DOI] [PubMed] [Google Scholar]
- Simons M., Mlodzik M. (2008). Planar cell polarity signaling: from fly development to human disease. Annu. Rev. Genet. 42, 517-540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiczka K. S., Yeaman C. (2008). Ral-regulated interaction between Sec5 and paxillin targets exocyst to focal complexes during cell migration. J. Cell Sci. 121, 2880-2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., Adachi A. (1998). Nuclear access and action of Notch in vivo. Cell 93, 649-660 [DOI] [PubMed] [Google Scholar]
- Strutt D., Johnson R., Cooper K., Bray S. (2002). Asymmetric localization of Frizzled and the determination of Notch-dependent cell fate in the Drosophila eye. Curr. Biol. 12, 813-824 [DOI] [PubMed] [Google Scholar]
- Strutt H., Strutt D. (2005). Long-range coordination of planar polarity in Drosophila. BioEssays 27, 1218-1227 [DOI] [PubMed] [Google Scholar]
- Strutt H., Strutt D. (2009). Asymmetric localization of planar polarity proteins: mechanisms and consequences. Semin. Cell Dev. Biol. 20, 957-963 [DOI] [PubMed] [Google Scholar]
- Sugihara K., Asano S., Tanaka K., Iwamatsu A., Okawa K., Ohta Y. (2002). The exocyst complex binds the small GTPase RalA to mediate filopodia. Nat. Cell Biol. 4, 73-78 [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Ready D. F. (1987). Neuronal differentiation in the Drosophila ommatidium. Dev. Biol. 120, 366-376 [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Struhl G. (1999). Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development 126, 5725-5738 [DOI] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., Marygold S., Millburn G., Osumi-Sutherland D., Shroeder A., Seal R., et al. (2009). FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37, D555-D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam E. M., Robinson P. J. (2006). Ral: mediator of membrane trafficking. Intl. J. Biochem. Cell Biol. 38, 1841-1847 [DOI] [PubMed] [Google Scholar]
- Wang W., Struhl G. (2004). Drosophila epsin mediates a select endocytic pathway that DSL ligands must enter to activation Notch. Development 131, 5367-5380 [DOI] [PubMed] [Google Scholar]
- Wang W., Struhl G. (2005). Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 132, 2883-2894 [DOI] [PubMed] [Google Scholar]
- Wilkin M. B., Tongngok P., Gensch N., Clemence S., Motoki M., Yamada K., Hori K., Taniguchi-Kanai M., Franklin E., Matsuno K., et al. (2008). Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of Notch in the endosomal trafficking pathway. Dev. Cell 15, 762-772 [DOI] [PubMed] [Google Scholar]
- Wolff T., Ready D. F. (1993). Pattern formation in the Drosophila retina. In The Development of Drosophila melanogaster, Vol. 2 (ed. Bate M., Martinez Arias A.), pp. 1277-1325 Cold Spring Harbor: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Wu H., Rossi G., Brennwald P. (2008). The ghost in the machine: small GTPases as spatial regulators of exocytosis. Trends Cell Biol. 18, 397-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Mlodzik M. (2009). A quest for the mechanism regulating global planar cell polarity of tissues. Trends Cell Biol. 19, 295-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Denef N., Schupbach T. (2009). The vacuolar proton pump, V-ATPase, is required for Notch signaling and endosomal trafficking in Drosophila. Dev. Cell 17, 387-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Zhang J., Carthew R. W. (1995). frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development 121, 3045-3055 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.