Abstract
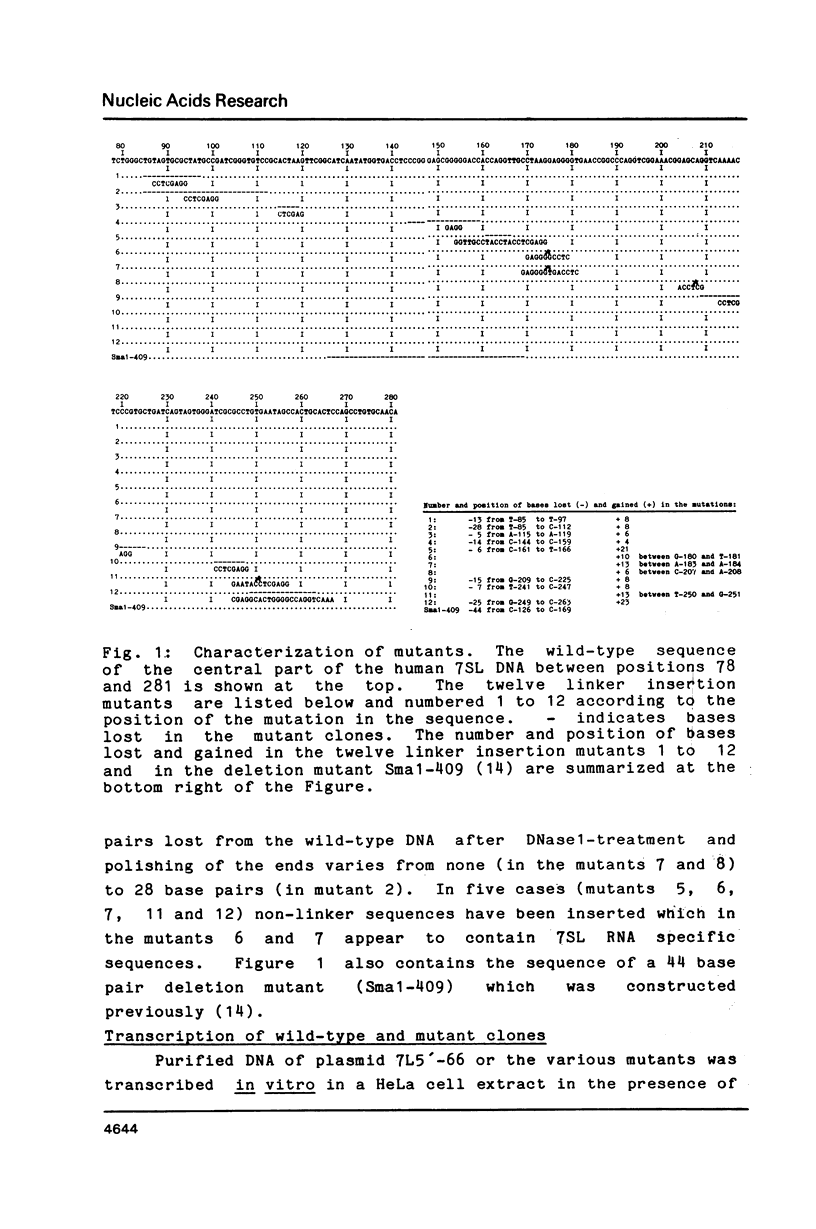
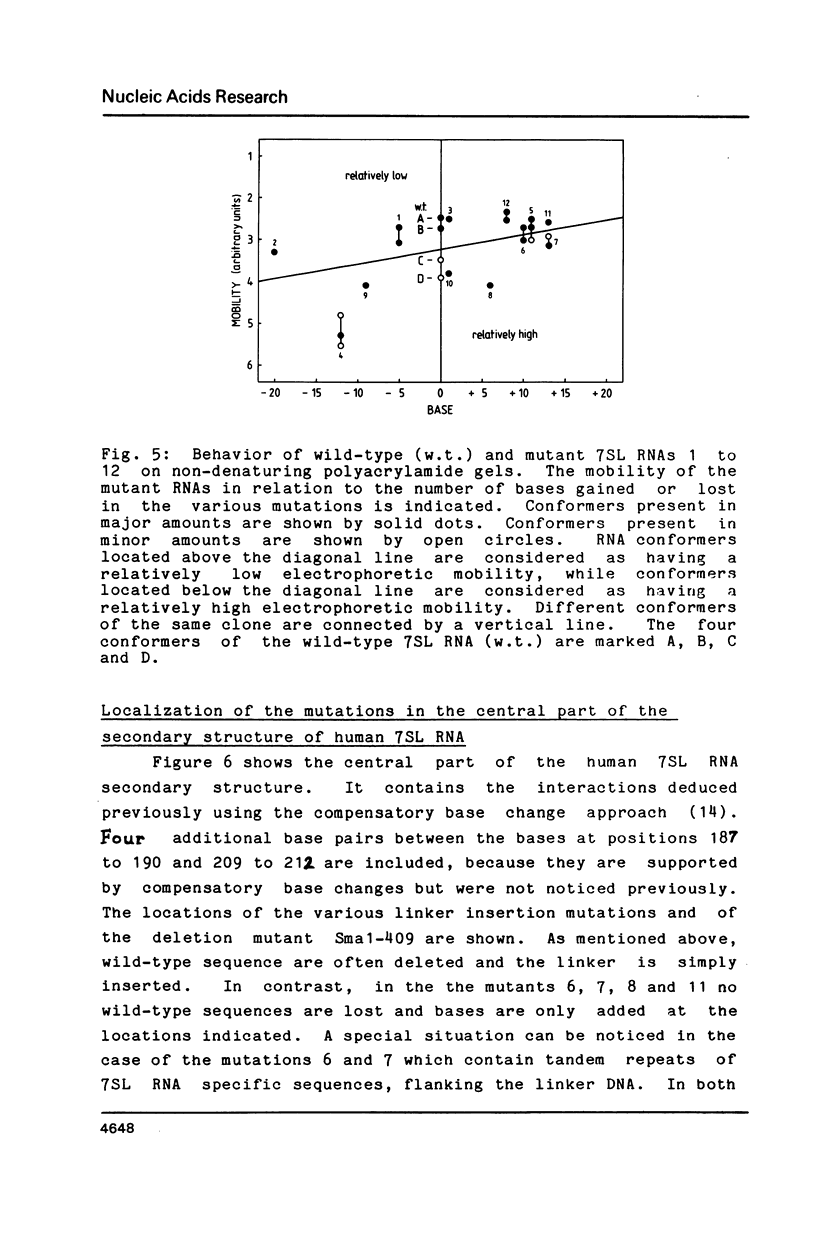
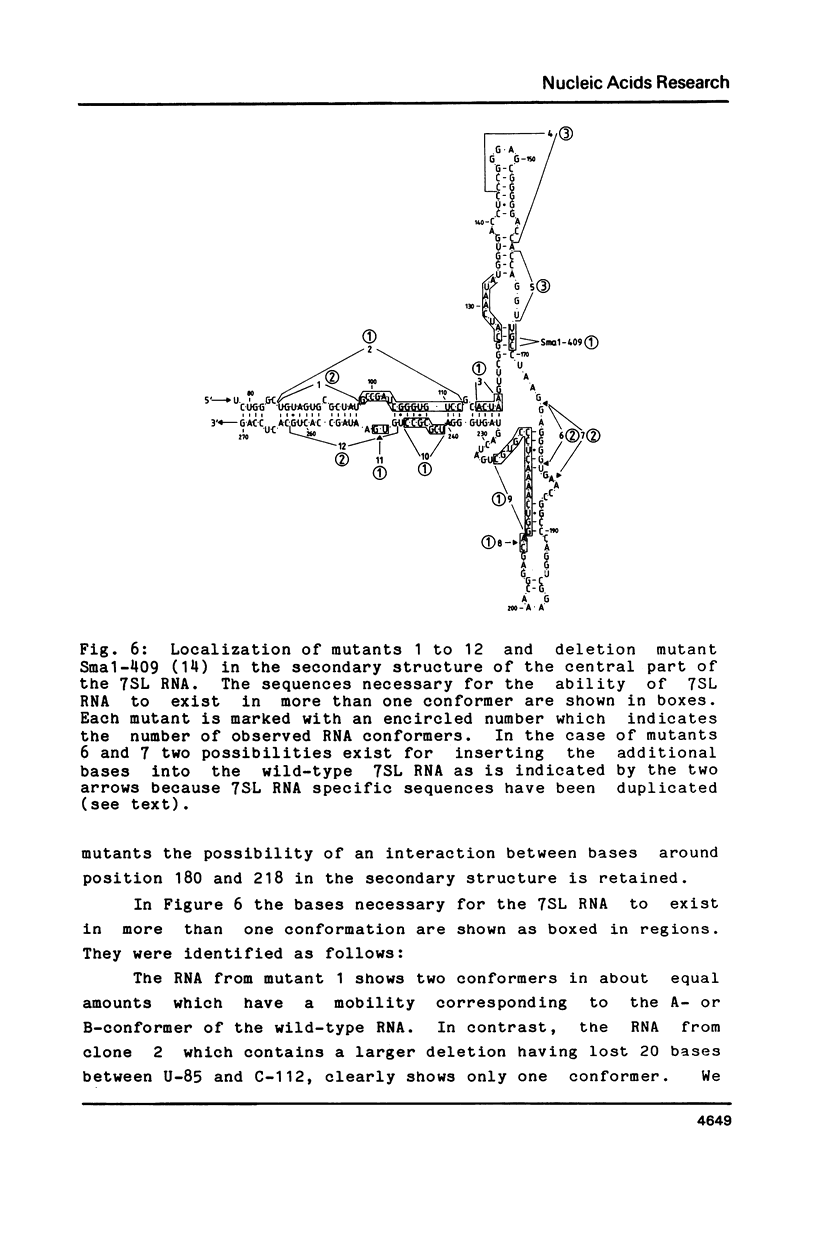
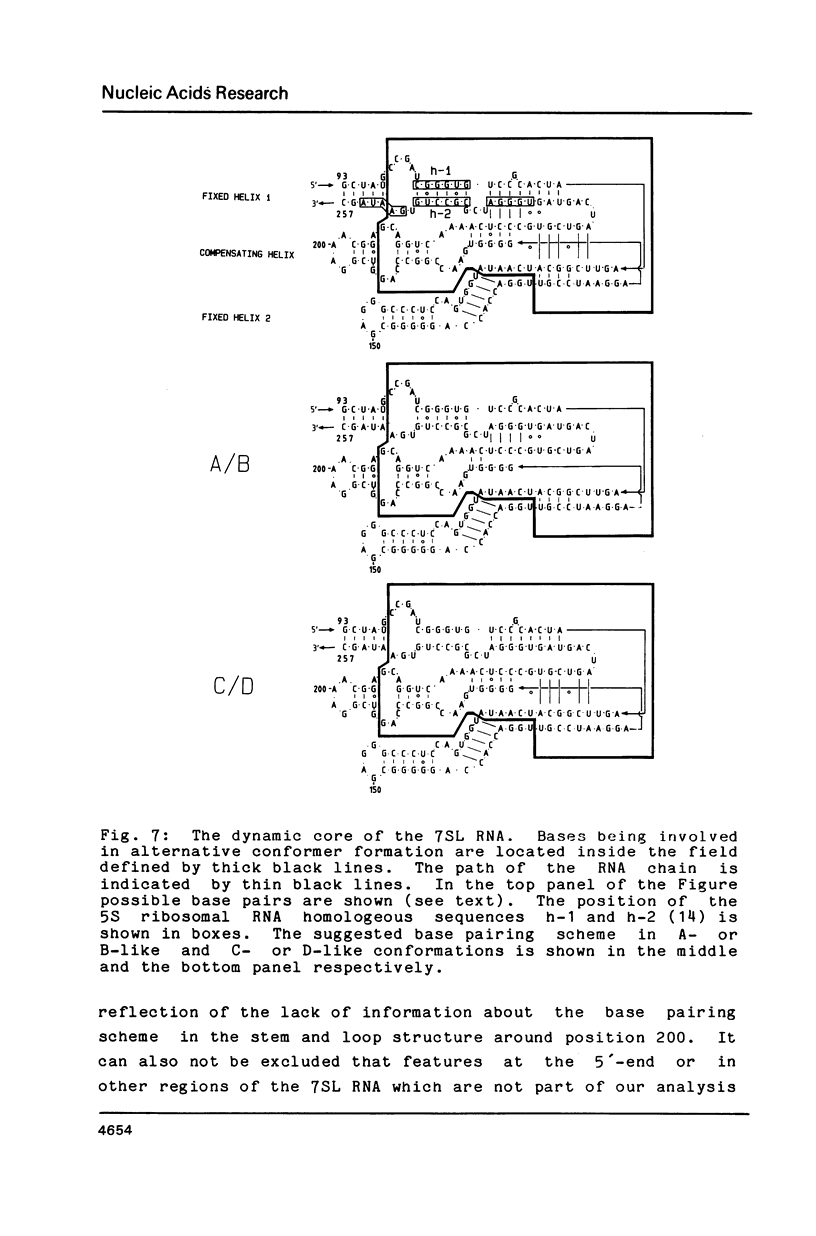
The human 7SL RNA component of the signal recognition particle can be separated into four major conformers by nondenaturing polyacrylamide gel electrophoresis. We have investigated what sequences give 7SL RNA the property to exist in different conformations. The human 7SL gene 7L30.1 was mutagenized using the random linker insertion approach and twelve mutant genes carrying alterations in the central domain of 7SL RNA were characterized. Mutant RNAs were produced by in vitro transcription of the various templates and their electrophoretic behaviour was determined. Bases between positions 98 and 133 as well as 206 and 251 proved to be necessary for the 7SL RNA to be able to exist in alternative conformations, while changes at the positions 85 to 97, 144 to 166 and 252 to 266 did not abolish this property. The dynamic sequences are located in the "central T" in the secondary structure of the 7SL RNA. They are phylogenetically conserved and include bases which are homologeous to 5S ribosomal RNA. A dynamic core structure composed of the dynamic parts of the 7SL RNA is suggested. An attempt was made to define the different conformers present in the wild-type 7SL RNA. These alternative configurations could play a functional role during the initial stage of protein translocation across the membrane of the endoplasmic reticulum.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brimacombe R., Maly P., Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. L., Gutell R., Noller H. F., Wool I. G. The nucleotide sequence of a rat 18 S ribosomal ribonucleic acid gene and a proposal for the secondary structure of 18 S ribosomal ribonucleic acid. J Biol Chem. 1984 Jan 10;259(1):224–230. [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Equilibria in 5-S ribosomal RNA secondary structure. Bulges and interior loops in 5-S RNA secondary structure may serve as articulations for a flexible molecule. Eur J Biochem. 1984 Aug 15;143(1):175–182. doi: 10.1111/j.1432-1033.1984.tb08356.x. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M., Rigler R., Wintermeyer W. On the structure and conformational dynamics of yeast phenylalanine-accepting transfer ribonucleic acid in solution. Biochemistry. 1979 Oct 16;18(21):4588–4599. doi: 10.1021/bi00588a020. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science. 1984 Jan 20;223(4633):285–286. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- Gundelfinger E. D., Di Carlo M., Zopf D., Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984 Oct;3(10):2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D., Krause E., Melli M., Dobberstein B. The organization of the 7SL RNA in the signal recognition particle. Nucleic Acids Res. 1983 Nov 11;11(21):7363–7374. doi: 10.1093/nar/11.21.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K., Cruz P., Tinoco I., Jr, Jovin T. M., van de Sande J. H. 'Z-RNA'--a left-handed RNA double helix. Nature. 1984 Oct 11;311(5986):584–586. doi: 10.1038/311584a0. [DOI] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese P., Symons R. H. Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4582–4586. doi: 10.1073/pnas.82.14.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B. K., Staden A., Schlessinger D. Alternative conformations in Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3539–3542. doi: 10.1073/pnas.82.11.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. A., Guertin D., Sonenberg N. mRNA secondary structure as a determinant in cap recognition and initiation complex formation. ATP-Mg2+ independent cross-linking of cap binding proteins to m7I-capped inosine-substituted reovirus mRNA. J Biol Chem. 1983 Jan 25;258(2):707–710. [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Ullu E., Weiner A. M. Human genes and pseudogenes for the 7SL RNA component of signal recognition particle. EMBO J. 1984 Dec 20;3(13):3303–3310. doi: 10.1002/j.1460-2075.1984.tb02294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Weiner A. M. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. 1985 Nov 28-Dec 4Nature. 318(6044):371–374. doi: 10.1038/318371a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Zwieb C. The secondary structure of the 7SL RNA in the signal recognition particle: functional implications. Nucleic Acids Res. 1985 Sep 11;13(17):6105–6124. doi: 10.1093/nar/13.17.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]




