Abstract
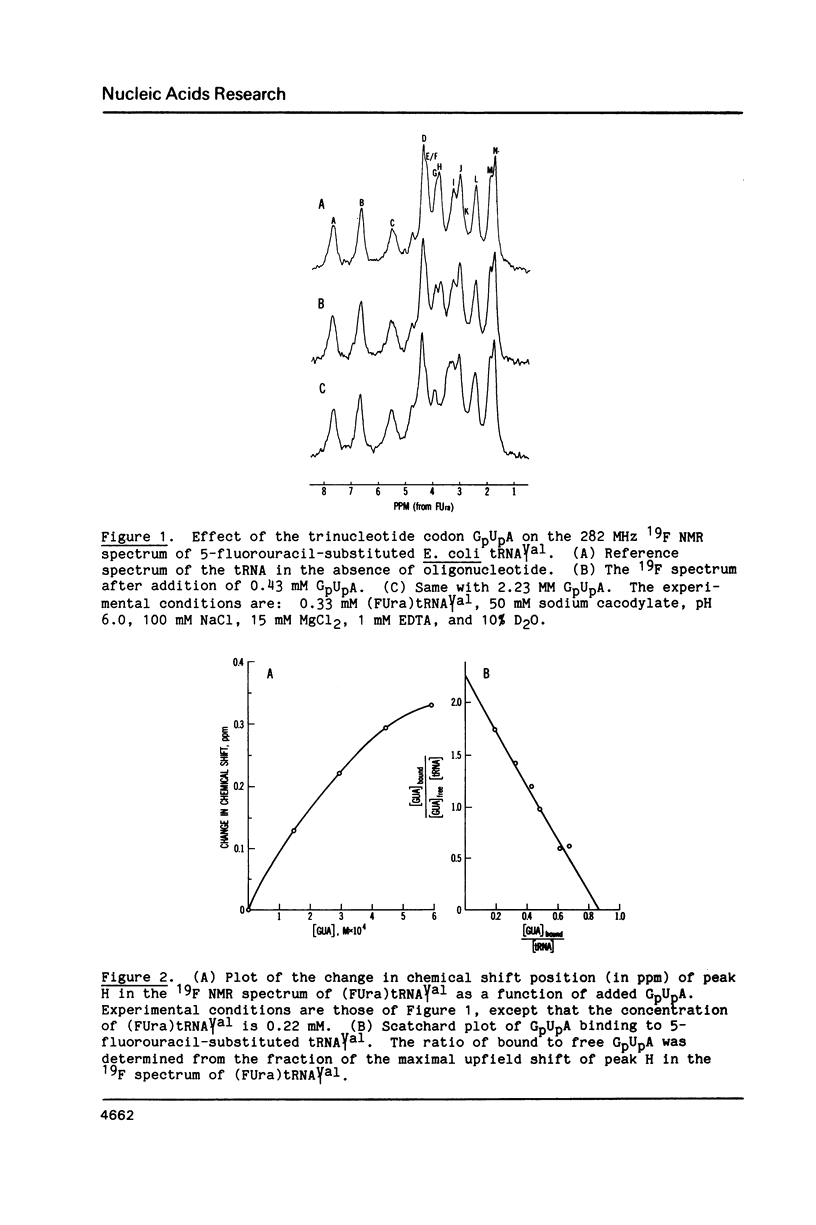
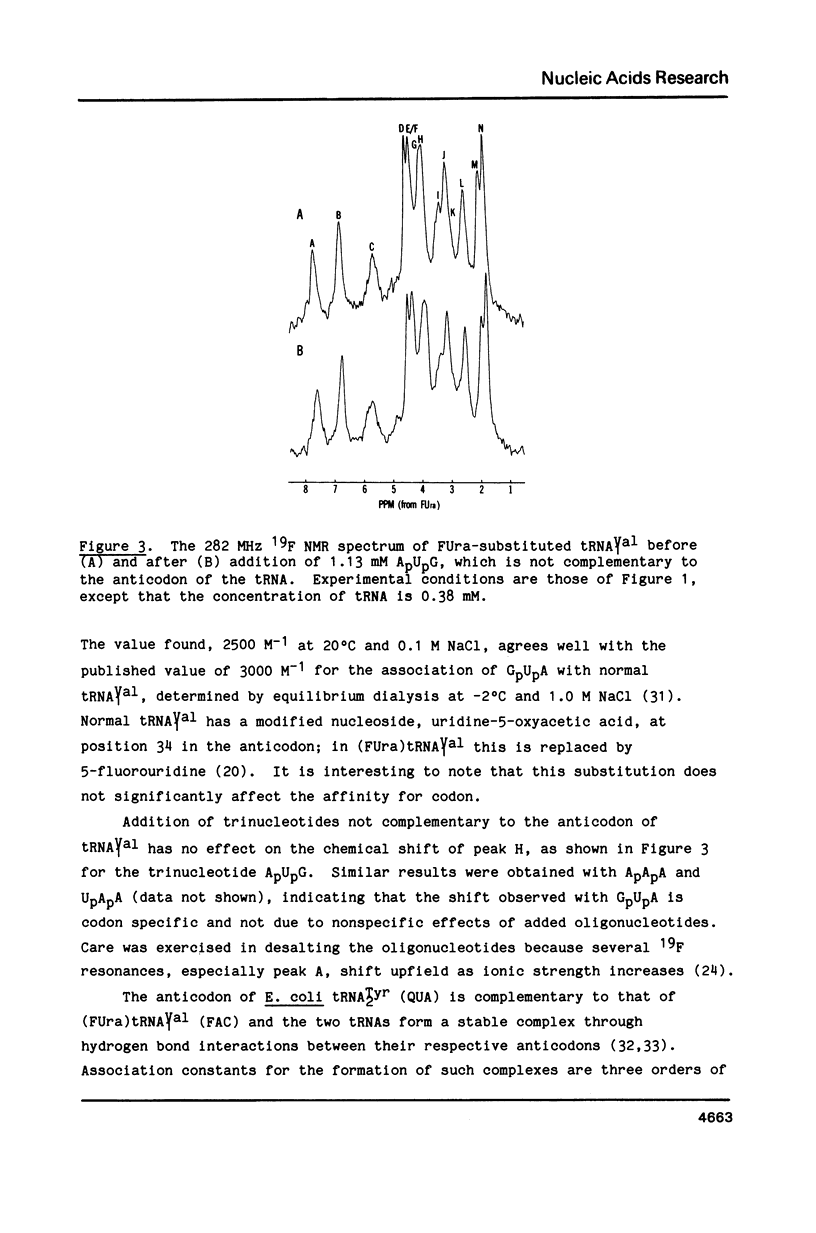
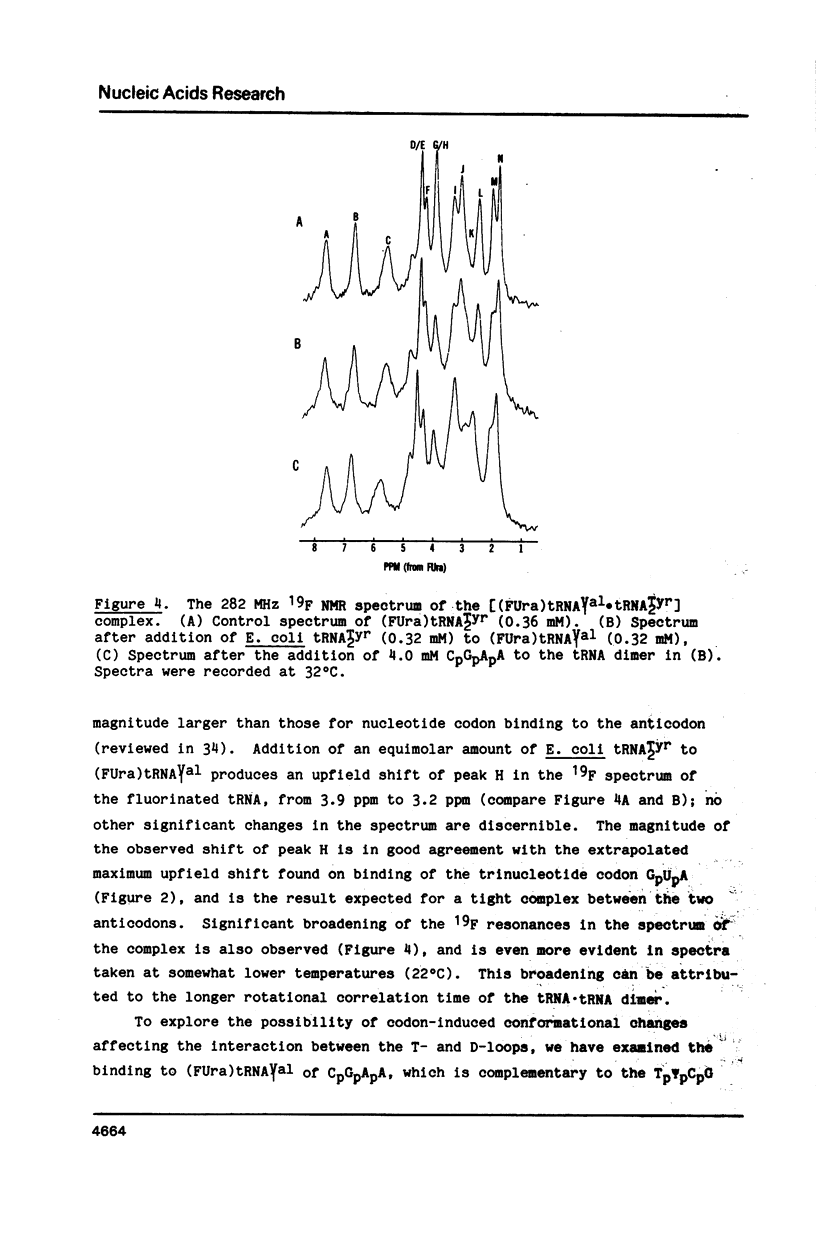
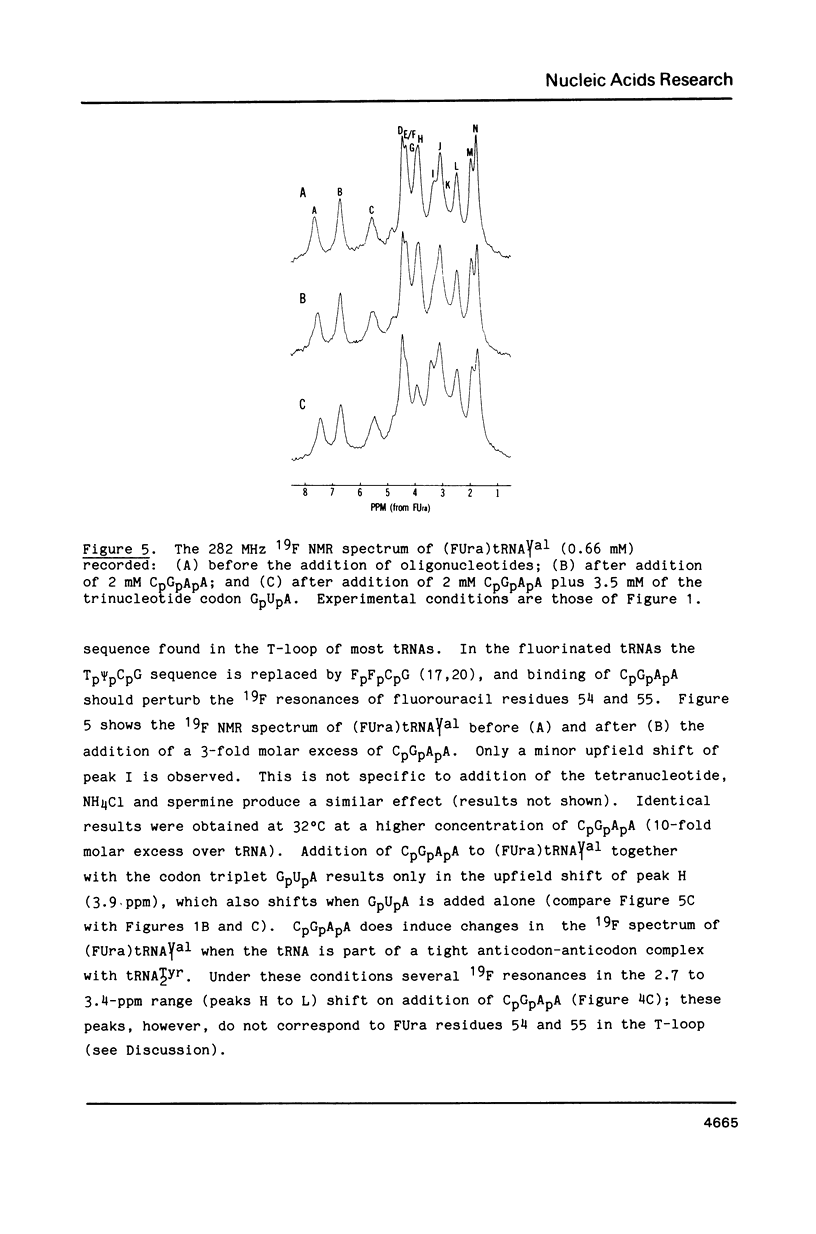
Codon-anticodon interaction was investigated in fully active 5-fluorouracil-substituted E. coli tRNAVal1 (anticodon FAC) by 19F NMR spectroscopy. Binding of the codon GpUpA results in the upfield shift of a 19F resonance at 3.9 ppm in the central region of the 19F NMR spectrum, whereas trinucleotides not complementary to the anticodon have no effect. The same 19F resonance shifts upfield upon formation of an anticodon-anticodon dimer between the 19F-labeled tRNA and E. coli tRNATyr2 (anticodon QUA). These results permit assignment of the peak at 3.9 ppm to the 5-fluorouracil at position 34 in the anticodon of fluorouracil-substituted tRNAVal1. The methionine codon ApUpG also causes a sequence-specific upfield shift of a peak in the central part of the 19F NMR spectrum of fluorinated E. coli tRNAMetm. However, ApUpG has no effect on the 19F spectrum of 19F-labeled E. coli tRNAMetf, indicating possible conformational differences between the anticodon loop of initiator and chain-elongating methionine tRNAs. 19F NMR experiments detect no binding of CpGpApA to the complementary FpFpCpG (replaces Tp psi pCpG) in the T-loop of 5-fluorouracil-substituted tRNAVal1, in the presence or absence of codon, suggesting that the tertiary interactions between the T- and D-loops are not disrupted by codon-anticodon interactions.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chinali G., Horowitz J., Ofengand J. Replacement of pseudouridine in transfer RNA by 5-fluorouridine does not affect the ability to stimulate the synthesis of guanosine 5'-triphosphate 3'-diphosphate. Biochemistry. 1978 Jul 11;17(14):2755–2760. doi: 10.1021/bi00607a009. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., McLaughlin L. W. Structure of the ribotrinucleoside diphosphate codon UpUpC bound to tRNAPhe from yeast. A time-dependent transferred nuclear Overhauser enhancement study. J Mol Biol. 1984 Mar 25;174(1):163–173. doi: 10.1016/0022-2836(84)90370-x. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Sprinzl M., Cramer F. Proton nuclear magnetic resonance of minor nucleosides in yeast phenylalanine transfer ribonucleic acid. Conformational changes as a consequence of aminoacylation, removal of the Y base, and codon--anticodon interaction. Biochemistry. 1979 Jul 24;18(15):3189–3199. doi: 10.1021/bi00582a001. [DOI] [PubMed] [Google Scholar]
- Eisinger J., Gross N. The anticodon-anticodon complex. J Mol Biol. 1974 Sep 5;88(1):165–174. doi: 10.1016/0022-2836(74)90302-7. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Tinoco I., Jr The binding of complementary oligoribonucleotides to yeast initiator Transfer RNA. Biochemistry. 1975 Jul 29;14(15):3310–3314. doi: 10.1021/bi00686a004. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Wataya Y., Hayatsu H., Ukita T. Chemical modification of tRNA-Tyr-yeast with bisulfite. A new method to modify isopentenyladenosine residue. Biochem Biophys Res Commun. 1970 Dec 9;41(5):1185–1191. doi: 10.1016/0006-291x(70)90211-1. [DOI] [PubMed] [Google Scholar]
- Gassen H. G. Ligand-induced conformational changes in ribonucleic acids. Prog Nucleic Acid Res Mol Biol. 1980;24:57–86. doi: 10.1016/s0079-6603(08)60671-6. [DOI] [PubMed] [Google Scholar]
- Geerdes H. A., Van Boom J. H., Hilbers C. W. Codon-anticodon interaction in tRNAPhe. II. A nuclear magnetic resonance study of the binding of the codon UUC. J Mol Biol. 1980 Sep 15;142(2):219–230. doi: 10.1016/0022-2836(80)90046-7. [DOI] [PubMed] [Google Scholar]
- Geerdes H. A., Van Boom J. H., Hilbers C. W. Nuclear magnetic resonance studies of codon-anticodon interaction in tRNAPhe. I. Effect of binding complementary tetra and pentanucleotides to the anticodon. J Mol Biol. 1980 Sep 15;142(2):195–217. doi: 10.1016/0022-2836(80)90045-5. [DOI] [PubMed] [Google Scholar]
- Geerdes H. A., van Boom J. H., Hilbers C. W. Codon--anticodon interaction in yeast tRNAPhe: an 1H NMR study. FEBS Lett. 1978 Apr 1;88(1):27–32. doi: 10.1016/0014-5793(78)80599-7. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Söll D. G., Crothers D. M. Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J Mol Biol. 1976 May 25;103(3):499–519. doi: 10.1016/0022-2836(76)90214-x. [DOI] [PubMed] [Google Scholar]
- Hills D. C., Cotten M. L., Horowitz J. Isolation and characterization of two 5-fluorouracil-substituted Escherichia coli initiator methionine transfer ribonucleic acids. Biochemistry. 1983 Mar 1;22(5):1113–1122. doi: 10.1021/bi00274a019. [DOI] [PubMed] [Google Scholar]
- Horowitz J., Ofengand J., Daniel W. E., Jr, Cohn M. 19F nuclear magnetic resonance of 5-fluorouridine-substituted tRNA1Val from Escherichia coli. J Biol Chem. 1977 Jun 25;252(12):4418–4420. [PubMed] [Google Scholar]
- Horowitz J., Ou C. N., Ishaq M. Isolation and partial characterization of Escherichia coli valine transfer RNA with uridine-derived residues replaced by 5-fluorouridine. J Mol Biol. 1974 Sep 15;88(2):301–312. doi: 10.1016/0022-2836(74)90483-5. [DOI] [PubMed] [Google Scholar]
- Högenauer G., Turnowsky F., Unger F. M. Codon-anticodon interaction of methionine specific tRNAs. Biochem Biophys Res Commun. 1972 Mar 24;46(6):2100–2106. doi: 10.1016/0006-291x(72)90765-6. [DOI] [PubMed] [Google Scholar]
- Labuda D., Pörschke D. Codon-induced transfer RNA association. A property of transfer RNA involved in its adaptor function? J Mol Biol. 1983 Jun 15;167(1):205–209. doi: 10.1016/s0022-2836(83)80042-4. [DOI] [PubMed] [Google Scholar]
- Labuda D., Striker G., Porschke D. Mechanism of codon recognition by transfer RNA and codon-induced tRNA association. J Mol Biol. 1984 Apr 25;174(4):587–604. doi: 10.1016/0022-2836(84)90085-8. [DOI] [PubMed] [Google Scholar]
- Mohr S. C., Thach R. E. Application of ribonuclease T1 to the synthesis of oligoribonucleotides of defined base sequence. J Biol Chem. 1969 Dec 25;244(24):6566–6576. [PubMed] [Google Scholar]
- Ofengand J., Bierbaum J. Protein synthetic ability of Escherichia coli valine transfer RNA with pseudouridine, ribothymidine, and other uridine-derived residues replaced by 5-fluorouridine. J Mol Biol. 1974 Sep 15;88(2):313–325. doi: 10.1016/0022-2836(74)90484-7. [DOI] [PubMed] [Google Scholar]
- Pongs O., Bald R., Reinwald E. On the structure of yeast tRNA Phe . Complementary-oligonucleotide binding studies. Eur J Biochem. 1973 Jan 3;32(1):117–125. doi: 10.1111/j.1432-1033.1973.tb02586.x. [DOI] [PubMed] [Google Scholar]
- Pongs O., Griese K. Complementary timer binding to transfer-RNA Val 1 . FEBS Lett. 1972 Oct 1;26(1):297–300. doi: 10.1016/0014-5793(72)80597-0. [DOI] [PubMed] [Google Scholar]
- Pörschke D., Labuda D. Codon-induced transfer ribonucleic acid association: quantitative analysis by sedimentation equilibrium. Biochemistry. 1982 Jan 5;21(1):53–56. doi: 10.1021/bi00530a010. [DOI] [PubMed] [Google Scholar]
- Ramberg E. S., Ishaq M., Rulf S., Moeller B., Horowitz J. Inhibition of transfer RNA function by replacement of uridine and uridine-derived nucleosides with 5-fluorouridine. Biochemistry. 1978 Sep 19;17(19):3978–3985. doi: 10.1021/bi00612a016. [DOI] [PubMed] [Google Scholar]
- Randerath K., Gupta R. C., Randerath E. 3H and 32P derivative methods for base composition and sequence analysis of RNA. Methods Enzymol. 1980;65(1):638–680. doi: 10.1016/s0076-6879(80)65065-4. [DOI] [PubMed] [Google Scholar]
- Schetters H., Gassen H. G., Matthaei H. Codon-anticodon interaction studied with oligonucleotides containing 3 -deazauridine, 4 -deoxyuridine or 3 -deaza- 4 -deoxyuridine. I. Synthesis by primer-dependent polynucleotide phosphorylase of oligonucleotides containing modofied nucleosides. Biochim Biophys Acta. 1972 Jul 31;272(4):549–559. doi: 10.1016/0005-2787(72)90510-2. [DOI] [PubMed] [Google Scholar]
- Schwarz U., Menzel H. M., Gassen H. G. Codon-dependent rearrangement of the three-dimensional structure of phenylalanine tRNA, exposing the T-psi-C-G sequence for binding to the 50S ribosomal subunit. Biochemistry. 1976 Jun 1;15(11):2484–2490. doi: 10.1021/bi00656a035. [DOI] [PubMed] [Google Scholar]
- THACH R. E., DOTY P. ENZYMATIC SYNTHESIS OF TRI- AND TETRANUCLEOTIDES OF DEFINED SEQUENCE. Science. 1965 Apr 30;148(3670):632–634. doi: 10.1126/science.148.3670.632. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. Complementary oligonucleotide binding to transfer RNA. J Mol Biol. 1972 Mar 14;65(1):25–41. doi: 10.1016/0022-2836(72)90489-5. [DOI] [PubMed] [Google Scholar]
- Wagner R., Garrett R. A. Chemical evidence for a codon-induced allosteric change in tRNALys involving the 7-methylguanosine residue 46. Eur J Biochem. 1979 Jul;97(2):615–621. doi: 10.1111/j.1432-1033.1979.tb13151.x. [DOI] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Wrede P., Woo N. H., Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3289–3293. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]


