Abstract
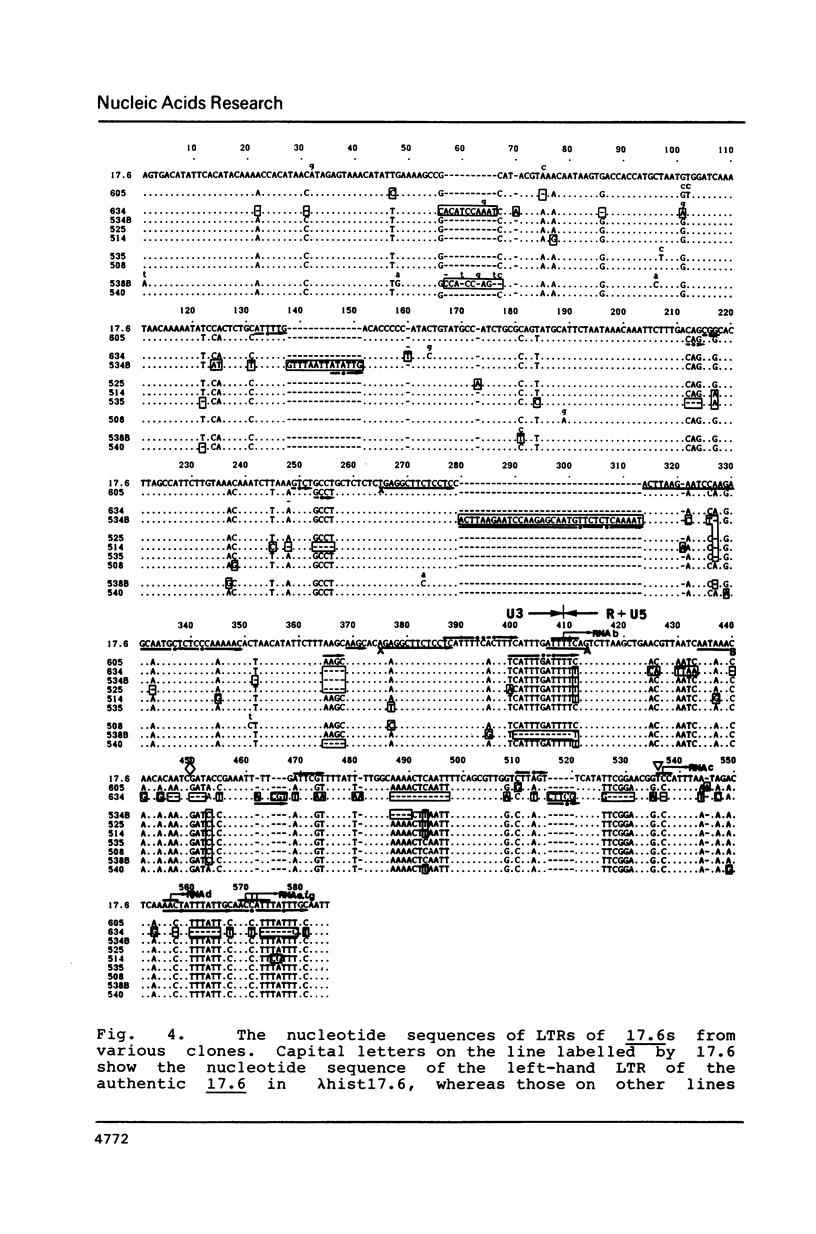
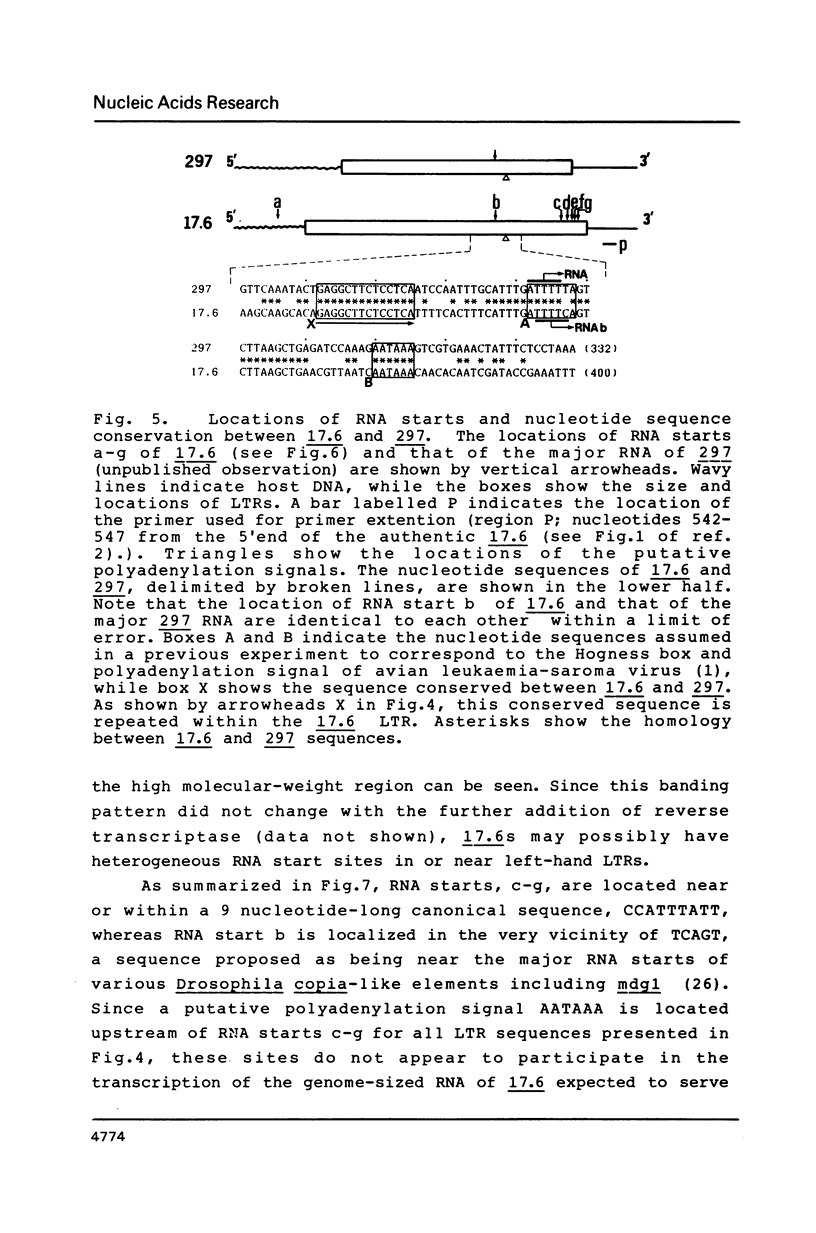
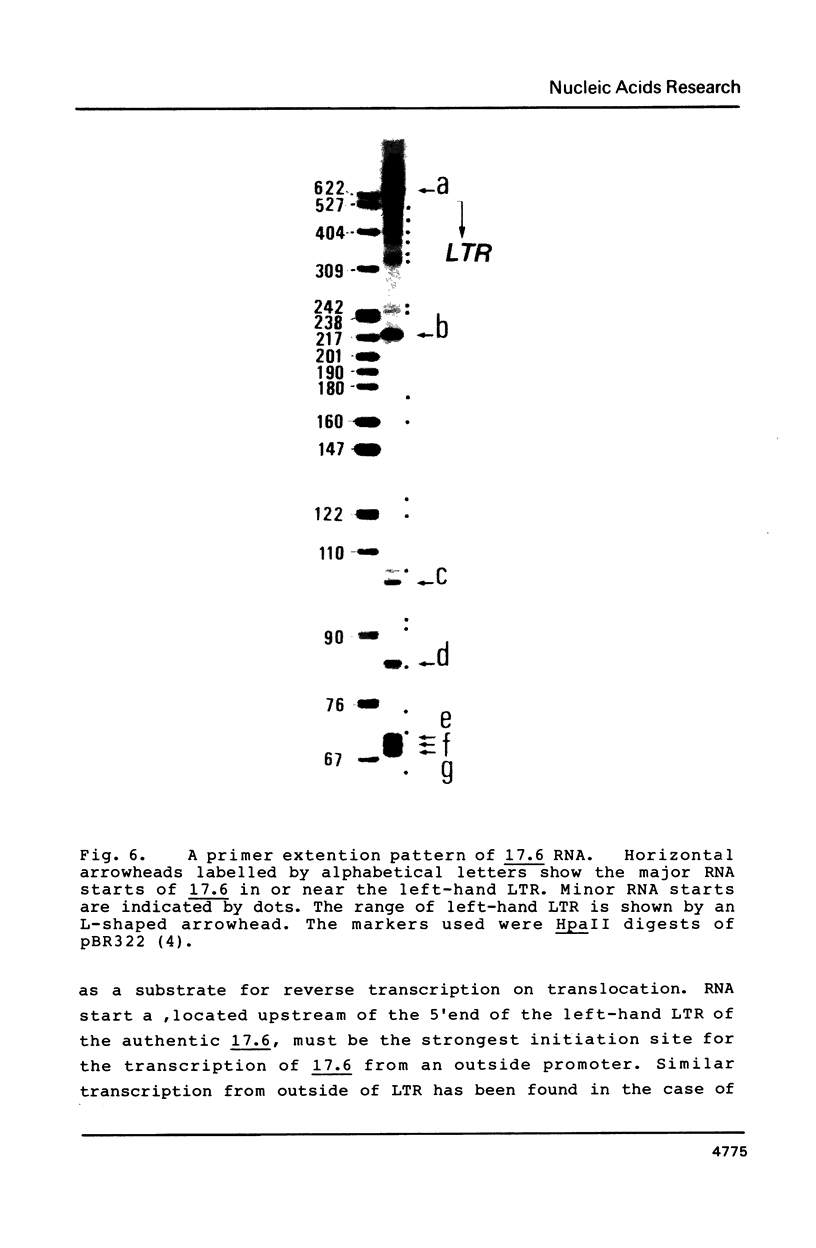
More than 21 members of 17.6, a Drosophila retrotransposon, were isolated and their possible structural changes were examined by restriction mapping, blot hybridization, heteroduplex analysis and nucleotide sequence determination of long terminal repeats (LTRs). At least 7 members were found to suffer with terminal or internal long deletions. No pair of LTRs having an identical nucleotide sequence was found either within an element or between elements. Although an initiation site for the presumable genome-sized transcript of 17.6, a potential substrate for reverse transcription on translocation, was identified within the left-hand LTR, our results as a whole support the notion that the majority of 17.6s have continued to reside for a long period of time at their present chromosomal loci and hence the rate of translocation of 17.6 is very low.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arkhipova I. R., Mazo A. M., Cherkasova V. A., Gorelova T. V., Schuppe N. G., Llyin Y. V. The steps of reverse transcription of Drosophila mobile dispersed genetic elements and U3-R-U5 structure of their LTRs. Cell. 1986 Feb 28;44(4):555–563. doi: 10.1016/0092-8674(86)90265-5. [DOI] [PubMed] [Google Scholar]
- Bayev A. A., Jr, Lyubomirskaya N. V., Dzhumagaliev E. B., Ananiev E. V., Amiantova I. G., Ilyin Y. V. Structural organization of transposable element mdg4 from Drosophila melanogaster and a nucleotide sequence of its long terminal repeats. Nucleic Acids Res. 1984 Apr 25;12(8):3707–3723. doi: 10.1093/nar/12.8.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley R. G., Gordon J. I., Burns A. T., Mullinix K. P., Binastein M., Goldberg R. F. Primary activation of the vitellogenin gene in the rooster. J Biol Chem. 1977 Nov 25;252(22):8310–8319. [PubMed] [Google Scholar]
- Emori Y., Shiba T., Kanaya S., Inouye S., Yuki S., Saigo K. The nucleotide sequences of copia and copia-related RNA in Drosophila virus-like particles. 1985 Jun 27-Jul 3Nature. 315(6022):773–776. doi: 10.1038/315773a0. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Levis R., Simon M. A., Rubin G. M. The 5' termini of RNAs encoded by the transposable element copia. Nucleic Acids Res. 1981 Dec 11;9(23):6279–6291. doi: 10.1093/nar/9.23.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund R., Meselson M. Long terminal repeat nucleotide sequence and specific insertion of the gypsy transposon. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4462–4464. doi: 10.1073/pnas.81.14.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Nelböck-Hochstetter P., Feldmann H. Nucleotide sequence and characteristics of a Ty element from yeast. Nucleic Acids Res. 1985 Apr 25;13(8):2745–2758. doi: 10.1093/nar/13.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Yuki S., Saigo K. Complete nucleotide sequence and genome organization of a Drosophila transposable genetic element, 297. Eur J Biochem. 1986 Jan 15;154(2):417–425. doi: 10.1111/j.1432-1033.1986.tb09414.x. [DOI] [PubMed] [Google Scholar]
- Inouye S., Yuki S., Saigo K. Sequence-specific insertion of the Drosophila transposable genetic element 17.6. 1984 Jul 26-Aug 1Nature. 310(5975):332–333. doi: 10.1038/310332a0. [DOI] [PubMed] [Google Scholar]
- Kugimiya W., Ikenaga H., Saigo K. Close relationship between the long terminal repeats of avian leukosis-sarcoma virus and copia-like movable genetic elements of Drosophila. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3193–3197. doi: 10.1073/pnas.80.11.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980 Sep;21(2):581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Rubin G. M. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol. 1985 Jul;5(7):1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigo K., Kugimiya W., Matsuo Y., Inouye S., Yoshioka K., Yuki S. Identification of the coding sequence for a reverse transcriptase-like enzyme in a transposable genetic element in Drosophila melanogaster. Nature. 1984 Dec 13;312(5995):659–661. doi: 10.1038/312659a0. [DOI] [PubMed] [Google Scholar]
- Saigo K., Millstein L., Thomas C. A., Jr The organization of Drosophila melanogaster histone genes. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):815–827. doi: 10.1101/sqb.1981.045.01.100. [DOI] [PubMed] [Google Scholar]
- Shepherd B. M., Finnegan D. J. Structure of circular copies of the 412 transposable element present in Drosophila melanogaster tissue culture cells, and isolation of a free 412 long terminal repeat. J Mol Biol. 1984 Nov 25;180(1):21–40. doi: 10.1016/0022-2836(84)90428-5. [DOI] [PubMed] [Google Scholar]
- Shiba T., Saigo K. Retrovirus-like particles containing RNA homologous to the transposable element copia in Drosophila melanogaster. Nature. 1983 Mar 10;302(5904):119–124. doi: 10.1038/302119a0. [DOI] [PubMed] [Google Scholar]
- Toh H., Kikuno R., Hayashida H., Miyata T., Kugimiya W., Inouye S., Yuki S., Saigo K. Close structural resemblance between putative polymerase of a Drosophila transposable genetic element 17.6 and pol gene product of Moloney murine leukaemia virus. EMBO J. 1985 May;4(5):1267–1272. doi: 10.1002/j.1460-2075.1985.tb03771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Warmington J. R., Waring R. B., Newlon C. S., Indge K. J., Oliver S. G. Nucleotide sequence characterization of Ty 1-17, a class II transposon from yeast. Nucleic Acids Res. 1985 Sep 25;13(18):6679–6693. doi: 10.1093/nar/13.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B. M., Bayev A. A., Finnegan D. J. Nucleotide sequence of terminal repeats of 412 transposable elements of Drosophila melanogaster. A similarity to proviral long terminal repeats and its implications for the mechanism of transposition. J Mol Biol. 1981 Dec 25;153(4):897–915. doi: 10.1016/0022-2836(81)90458-7. [DOI] [PubMed] [Google Scholar]
- Yuki S., Ishimaru S., Inouye S., Saigo K. Identification of genes for reverse transcriptase-like enzymes in two Drosophila retrotransposons, 412 and gypsy; a rapid detection method of reverse transcriptase genes using YXDD box probes. Nucleic Acids Res. 1986 Apr 11;14(7):3017–3030. doi: 10.1093/nar/14.7.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]