Abstract
A novel D-galactosyl-β1–3-N-acetyl-D-hexosamine phosphorylase cloned from Bifidobacterium infantis (BiGalHexNAcP) was used with a recombinant E. coli K-12 galactokinase (GalK) for efficient one-pot two-enzyme synthesis of T antigens, galacto-N-biose (Galβ1–3GalNAc), lacto-N-biose (Galβ1–3GlcNAc), and their derivatives.
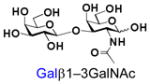
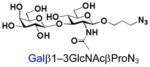
β1–3-Linked galactosides such as galacto-N-biose (GNB, Galβ1–3GalNAc) and lacto-N-biose (LNB, Galβ1–3GlcNAc) are important carbohydrate structures in nature. They are themselves, or are part of, important carbohydrate epitopes involved in cell adhesion, signalling, fertilization, differentiation, development, and cancer metastasis. For example, GNB with an α-configuration at the anomeric carbon of GalNAc (Galβ1–3GalNAcα) is named Thomsen-Friedenreich antigen (TF or T-antigen, Galβ1–3GalNAcαSer/Thr) disaccharide. It is the glycan of the Core 1 mucin-type O-GalNAc glycopeptides/glycoproteins and can be branched/extended to form Core 2 as well as extended Core 1 and Core 2 glycans.1 On the other hand, GNB with a β-configuration at the anomeric carbon of GalNAc (Galβ1–3GalNAcβ) is an essential part of the carbohydrate moieties of complex glycosphingolipids such as GA1, GM1, GD1, GT1, GQ1, and GP1, etc. In comparison, LNB is a common structure presented broadly in human milk oligosaccharides (e.g. lacto-N-tetraose and its sialylated/fucosylated derivatives) and in the carbohydrate moieties (e.g. type I glycans, Lewis a, sialyl Lewis a, and Lewis b antigens) of glycoproteins and glycolipids.1,2
To use these β1–3-linked galactosides as probes or glycan building blocks for biological studies, GNB and LNB have been synthesized by chemical3 and enzymatic approaches using β-glycosidases4 or β1–3-galactosyltransferases5. Nevertheless, these methods have limitations. Chemical synthesis requires multiple tedious protection-deprotection and purification procedures. Glycosidase-catalyzed reactions usually result in low yields and poor regioselectivity. The formation of β1–3-linked galactosidic bonds usingβ1–3-galactosyltransferases has been proven efficient, but its application is hampered by limited access to sufficient amount of highly efficient glycosyltransferases and the required sugar nucleotide, uridine 5′-diphosphate galactose (UDP-Gal).
Carbohydrate phosphorylases catalyze the reversible formation of monosaccharide-1-phosphate from an oligosaccharide or a polysaccharide and an inorganic phosphate. Their ability in catalyzing the reverse reaction has been used for the synthesis of oligosaccharides. Based on their protein sequence similarity, carbohydrate phosphorylases have been grouped into several families of glycoside hydrolases (GH13, GH65, GH94, GH112) and glycosyltransferases (GT3, GT4, GT20, GT35) in the Carbohydrate Active Enzymes (CAZy) database (http://www.cazy.org).6 Among them, D-galactosyl-β1–3-N-acetyl-D-hexosamine phosphorylase (GalHexNAcP, EC 2.4.1.211), a member of CAZy glycoside hydrolase GH112 family, was initially reported in Bifidobacterium bifidum DSM 20082 in 1999.7 It reversibly phosphorolyzes GNB and LNB to produce α-D-galactose-1-phosphate (Gal-1-P) and the corresponding N-acetyl-D-hexosamine. The partially purified enzyme (MW: 140 kDa)7 was used in the synthesis of Galβ1–3GlcNAcαOR containing different aglycones with low yields.8 The GalHexNAcP gene was later found in Clostridium perfringens ATCC13124, Vibrio vulnificus CMCP6, and several strains of bifidobacteria and was believed to contribute to the intestinal colonolization of the bacteria.9 The enzyme has been cloned and used with a sucrose phosphorylase, a UDP-glucose-hexose-1-phosphate uridylyltransferase, and a UDP-glucose 4-epimerase for one-pot four-enzyme high-yield large-scale production of LNB or GNB from sucrose and GlcNAc or GalNAc in the presence of UDP-glucose and phosphate.10
Crystal structure of a recombinant GalHexNAcP from Bifidobacterium longum JCM1217 reveals a partially broken TIM barrel fold resembling a thermophilic β-galactosidase.11 Nevertheless, the acceptor substrate specificity of GalHexNAcP has not been studied in detail and its application in efficient synthesis of GNB and LNB derivatives has not been fully explored. Here, we report a highly efficient one-pot two-enzyme approach for the synthesis of diverse β1–3-linked galactosides, including T antigens, GNB, LNB, and their derivatives using a novel D-galactosyl-β1–3-N-acetyl-D-hexosamine phosphorylase cloned from Bifidobacterium infantis (BiGalHexNAcP) and a recombinant galactokinase (GalK) cloned from E. coli K-12.12
To obtain BiGalHexNAcP, Blon_2174 gene (GenBank NC_011593) was amplified from the genomic DNA of Bifidobacterium longum subsp. infantis ATCC 1569713 by polymerase chain reaction and cloned into pET15b vector. Optimal expression of the recombinant N-His6-tagged protein (MW: 86.5 kDa) was achieved by incubating E. coli Origami™ B(DE3) cells containing desired plasmids at 25°C for 18–20 hours with vigorous shaking (250 rpm) after the addition of isopropyl-1-thio-β-D-galactopyranoside (IPTG) (0.1 mM). Ni2+-NTA column purification using an AKTA FPLC system yielded 55 mg pure protein per liter cell culture.
As shown in Scheme 1, the synthesis of GNB, LNB, and their derivatives was carried out using a one-pot two-enzyme system containing BiGalHexNAcP and a recombinant E. coli GalK. GalK catalyzed the formation of α-D-Gal-1-P from inexpensive D-Gal in the presence of ATP. The in situ generated α-D-Gal-1-P was used as the donor substrate by the phosphorylase to form diverseβ1–3-linked galactosides. Excess amounts of D-Gal and ATP (1.2 or 1.5 equiv.) were used to drive the reaction towards disaccharide formation.
Scheme 1.
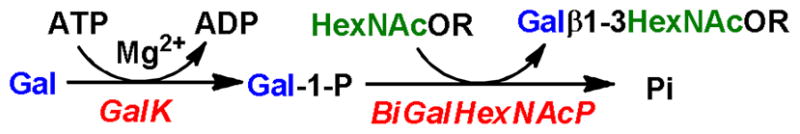
Synthetic scheme for one-pot two-enzyme synthesis of β1–3-linked galactosides. GalK, E. coli K-12 galactokinase; BiGalHexNAcP, Bifidobacterium infantis D-galactosyl-β1–3-N-acetyl-D-hexosamine phosphorylase.
HPLC-based pH profile study of BiGalHexNAcP revealed that its catalytic activity was optimum in a relatively narrow pH range of 5.0 to 6.5. Extremely low activity was observed when the pH of the reaction reached 7.0 or higher (see Supporting Information). For GalK, it prefered pH higher than 7.0 for optimum activity, showed medium activity at pH 6.5 and low activity at pH lower than 6.5. Therefore, pH 6.5 was chosen for the one-pot two-enzyme reactions. The reactions were carried out at 37°C for 48 hours. Products were purified using the combination of size exclusion chromatography and silica gel chromatography.
As shown in Table 1, BiGalHexNAcP exhibits promiscuous acceptor substrate specificity and has comparable levels of activity toward GlcNAc and GalNAc-based structures. For example, free GlcNAc (1) and GalNAc (11), their β-glycosides (2 and 12 respectively) and α-glycosides (3 and 13 respectively) are superb acceptors for BiGalHexNAcP to produce LNB (21–23) and GNB (31–33) disaccharides in exellent 92–96% yields. Apparently, the configurations of the C-4 hydroxyl group and the anomeric center of the N-acetyl hexosamine do not affect the activity of BiGalHexNAcP.
Table 1.
One-pot two-enzyme synthesis of β1–3-linked galactosides from Gal, ATP, and N-acetyl-hexosamine (HexNAc) derivatives using GalK and BiGalHexNAcP.
| Acceptors | Products | Yields (%) | Acceptors | Products | Yields (%) |
|---|---|---|---|---|---|
 1 |
 21 |
95 |
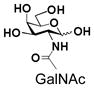 11 |
 31 |
93 |
 2 |
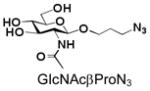
 22 |
96 |
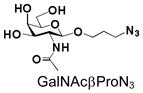 12 |
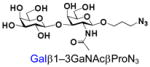 32 |
95 |
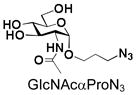 3 |
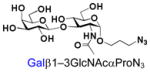 23 |
94 |
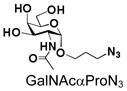 13 |
 33 |
92 |
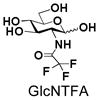 4 |
 24 |
93 |
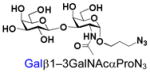
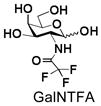 14 |
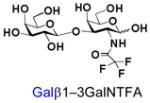 34 |
94 |
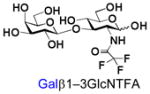
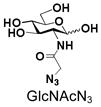 5 |
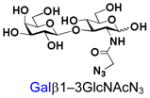 25 |
91 |
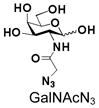 15 |
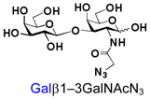 35 |
92 |
 6 |
 26 |
86 |
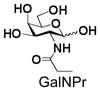 16 |
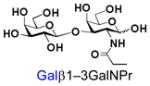 36 |
69 |
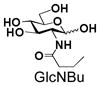 7 |
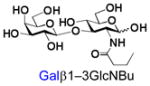 27 |
78 |
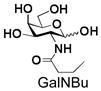 17 |
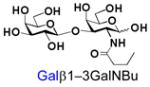 37 |
NR |
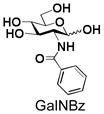 8 |
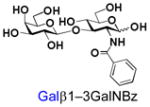 28 |
NR |
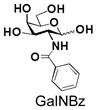 18 |
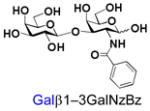 38 |
NR |
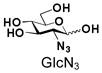 9 |
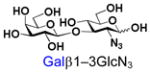 29 |
NR |
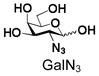 19 |
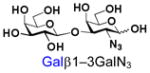 39 |
NR |
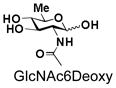 10 |
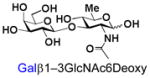 30 |
84 |
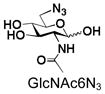 20 |
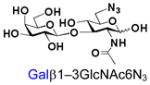 40 |
87 |
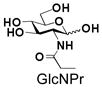
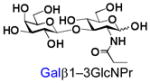
Similarly, N-acetyl group derivatization with small acyl groups or derivatives does not affect the BiGalHexNAcP activity signicantly. For example, GlcNTFA 4 and GalNTFA 14 with an N-trifluoroacetyl group or GlcNAcN3 5 and GalNAcN3 15 with an N-azidoacetyl group in the hexosamines are excellent acceptors to produce disaccharides 24–25 and 34–35 in 91–94% yields. However, bigger N-acyl groups differentiated glucosamine and galactosamine-based acceptors. For example, GlcNPr 6 and GlcNBu 7 with an N-propyl and an N-butyl group, respectively, at glucosamine are good acceptors for BiGalHexNAcP to synthesize disaccharides 26 and 27 in 86% and 78% yields, respectively. The synthetic yield decreases moderately as the size of the N-acyl group on the glucosamine increases. In contrast, the corresponding galactosamine derivative GalNPr 16 leads to disaccharide 36 in a moderate 69% yield while GalNBu 17 is not a suitable acceptor for BiGalHexNAcP. A bulky N-benzoyl group prevents both glucosamine and galactosamine derivatives (8 and 18) from being suitable BiGalHexNAcP acceptors. Quite interestingly, GlcN3 9 and GalN3 19, the C2 derivatives of GlcNAc and GalNAc with the N-acetyl group being replaced by a relatively small azido group (-N3), are not tolerable acceptors.
BiGalHexNAcP also shows good tolerance towards C6-modification. Both 6-deoxy-GlcNAc (GlcNAc6Deoxy) 10 and 6-azido-6-deoxy-GlcNAc (GlcNAc6N3) 20 are very good acceptors for the enzyme to synthesize disaccharides 30 and 40 in high yields (84 and 87%, respectively).
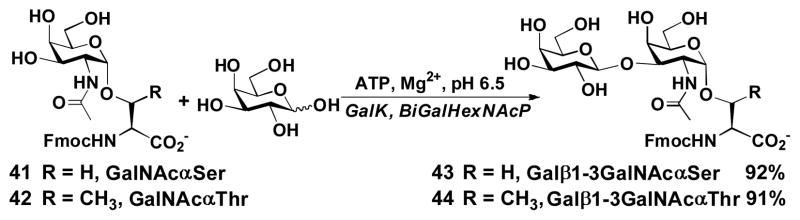
The BiGalHexNAcP and the one-pot two-enzyme system have also been successfully applied in the efficient synthesis of biologically important T antigens Galβ1–3GalNAcα1-O-Ser and Galβ1–3GalNAcα1-O-Thr. As shown in Scheme 2, incubating GalNAcα1-O-Ser 41 or GalNAcα1-O-Thr 42 with Gal and ATP in the presence of GalK and BiGalHexNAcP sucessfully produced the desired disaccharide products 43 and 44 in excellent 92% and 91% yields, respectively.
Scheme 2.
One-pot two-enzyme synthesis of T-antigen Galβ1–3GalNAcα1-O-Ser/Thr.
In summary, taking advantage of the acceptor substrate promiscuity of BiGalHexNAcP, we have developed a highly efficient one-pot two-enzyme approach for the synthesis of diverseβ1–3-linked galactosides. Compared to galactosyltransferase-catalyzed approaches, the BiGalHexNAcP-catalyzed reactions do not require the use, in situ generation, or regeneration of expensive sugar nucleotide, and thus are more efficient and simplified systems for producing galactosides. We believe that this synthetic route will contribute greatly to obtaining and elucidating the important roles of β1–3-galactosides as well as β1–3-galactoside-containing glycans and glycoconjugates.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01GM076360, U01CA128442 (to X. Chen), and R01HD061935 (to P.G. Wang). X. Chen is an Alfred P. Sloan Research Fellow, a Camille Dreyfus Teacher-Scholar, and a UC-Davis Chancellor’s Fellow. We thank Professor David Mills at the University of California-Davis for providing us the genomic DNA of Bifidobacterium longum subsp. infantis ATCC 15697.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details for cloning and characterization of BiGalHexNAcP, chemical and enzymatic synthesis, NMR and HRMS data. See DOI: 10.1039/b000000x/
Notes and references
- 1.Varki A, cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; New York: 2008. [PubMed] [Google Scholar]
- 2.(a) Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]; (b) Holgersson J, Lofling J. Glycobiology. 2006;16:584–593. doi: 10.1093/glycob/cwj090. [DOI] [PubMed] [Google Scholar]; (c) Feizi T. Immunol Rev. 2000;173:79–88. doi: 10.1034/j.1600-065x.2000.917310.x. [DOI] [PubMed] [Google Scholar]; (d) Bertozzi CR, Kiessling LL. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 3.Wilstermann M, Magnusson G. Carbohydr Res. 1995;272:1–7. doi: 10.1016/0008-6215(95)00026-p. [DOI] [PubMed] [Google Scholar]
- 4.(a) Hedbys L, Johansson E, Mosbach K, Larsson PO. Carbohydr Res. 1989;186:217–223. doi: 10.1016/0008-6215(89)84036-4. [DOI] [PubMed] [Google Scholar]; (b) Vetere A, Miletich M, Bosco M, Paoletti S. Eur J Biochem. 2000;267:942–949. doi: 10.1046/j.1432-1327.2000.01068.x. [DOI] [PubMed] [Google Scholar]; (c) D’Almeida A, Ionata M, Tran V, Tellier C, Dion M, Rabiller C. Tetrahedron-Asymmetry. 2009;20:1243–1246. [Google Scholar]
- 5.(a) Baisch G, Ohrlein R, Streiff M, Kolbinger F. Bioorg Med Chem Lett. 1998;8:751–754. doi: 10.1016/s0960-894x(98)00093-6. [DOI] [PubMed] [Google Scholar]; (b) Su DM, Eguchi H, Yi W, Li L, Wang PG, Xia C. Org Lett. 2008;10:1009–1012. doi: 10.1021/ol703121h. [DOI] [PubMed] [Google Scholar]; (c) Wang Z, Gilbert M, Eguchi H, Yu H, Cheng J, Muthana S, Zhou L, Wang PG, Chen X, Huang X. Adv Synth Catal. 2008;350:1717–1728. doi: 10.1002/adsc.200800129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derensy-Dron D, Krzewinski F, Brassart C, Bouquelet S. Biotechnol Appl Biochem. 1999;29(Pt 1):3–10. [PubMed] [Google Scholar]
- 8.Farkas E, Thiem J, Krzewinski F, Bouquelet S. Synlett. 2000:728–730. [Google Scholar]
- 9.(a) Kitaoka M, Tian J, Nishimoto M. Appl Environ Microbiol. 2005;71:3158–3162. doi: 10.1128/AEM.71.6.3158-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xiao J, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, Iwatsuki K, Kitaoka M. Appl Environ Microbiol. 2010;76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Nishimoto M, Kitaoka M. Biosci Biotechnol Biochem. 2007;71:2101–2104. doi: 10.1271/bbb.70320. [DOI] [PubMed] [Google Scholar]; (b) Nishimoto M, Kitaoka M. Carbohydr Res. 2009;344:2573–2576. doi: 10.1016/j.carres.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 11.Hidaka M, Nishimoto M, Kitaoka M, Wakagi T, Shoun H, Fushinobu S. J Biol Chem. 2009;284:7273–7283. doi: 10.1074/jbc.M808525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Fang J, Zhang J, Liu Z, Shao J, Kowal P, Andreana P, Wang PG. J Am Chem Soc. 2001;123:2081–2082. doi: 10.1021/ja005738v. [DOI] [PubMed] [Google Scholar]
- 13.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. Proc Natl Aca Sci U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.