Abstract
Despite the importance of neuraminidase (NA) activity in effective infection by influenza A viruses, limited information exists about the differences of substrate preferences of viral neuraminidases from different hosts or from different strains. Using a high-throughput screening format and a library of twenty α2–3- or α2–6-linked para-nitrophenol-tagged sialylgalactosides, substrate specificity of NAs on thirty-seven strains of human and avian influenza A viruses was studied using intact viral particles. Neuraminidases of all viruses tested cleaved both α2–3- and α2–6-linked sialosides but preferred α2–3-linked ones and the activity was dependent on the terminal sialic acid structure. In contrast to NAs of other subtypes of influenza A viruses which did not cleave 2-keto-3-deoxy-D-glycero-D-galacto-nonulosonic acid (Kdn) or 5-deoxy Kdn (5d–Kdn), NAs of all N7 subtype viruses tested had noticeable hydrolytic activities on α2–3-linked sialosides containing Kdn or 5d–Kdn. Additionally, group 1 NAs showed efficient activity in cleaving N-azidoacetylneuraminic acid from α2 –3-linked sialoside.
Keywords: Carbohydrate, Influenza A virus, Inhibitors, Neuraminidase, Sialic acid, Sialosides, Substrate specificity studies
Introduction
Human influenza A virus is an important pathogen that causes an acute viral disease of the respiratory tract in millions of people each year. Avian influenza A virus outbreaks have caused major losses for the poultry industry (Spicuzza et al., 2007), but can also cause deadly diseases (e.g. H5N1) as well as less severe infections in humans (Swayne and Halvorson, 2008). The 2009 pandemic by an influenza A virus strain of mixed avian and swine origins have stimulated renewed interest into emerging influenza virus strains.
Influenza A virions contain two types of surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), both recognizing sialic acid-containing receptors on the host cell surface. HAs bind to cell surface sialyloligosaccharides and mediate the entry of the virus into the cell (Wiley and Skehel, 1987). NAs are believed to prevent the aggregation of progeny virions by removing sialic acid from the carbohydrate moieties of newly synthesized HA and NA glycopolypeptides (Palese et al., 1974) and facilitate the spreading of progeny virions by removal of sialic acid from the glycoconjugates on the surface of infected host cells (von Itzstein, 2007). Recent studies found that in addition to its previously recognized functions, NA plays a direct and early role in influenza A virus entry into host target cells. The presence of NA significantly enhanced both pseudotype infectivity and cell-cell fusion (Su et al., 2009). Therefore, understanding the substrate specificity of influenza A virus NAs is critical for preventing direct transmission of virus from avian or other species to human and for developing new potent NA inhibitors.
Neuraminidases (EC 3.2.1.18) are sialoside hydrolyzing enzymes which cleave the glycosidic linkages between the terminal sialic acid and an adjacent sugar residue. To date, studies on the substrate specificity of influenza A virus neuraminidases are limited to a small number of influenza strains and a restricted set of sialosides. The NAs of N2 influenza A viruses isolated from different hosts were shown to differ in their ability to distinguish α2–3-linked and α2–6-linked sialyl lactose (Baum and Paulson, 1991; Couceiro and Baum, 1994; Franca de Barros et al., 2003; Kobasa et al., 1999). The recognition of underlying glycan structures of sialosides by human influenza A viruses has been observed (Katinger et al., 2004). More recently, the substrate specificity of the neuraminidases of H1N1 influenza A virus and avian-human reassortant influenza A viruses were studied using six sialosides, including BODIPY-labeled α2–3-linked sialyl lactose, α2–3-linked sialyl N-acetyllactosamine, sialyl Lewis c, sialyl Lewis a, α2–6-linked sialyl lactose, and α2–6-linked sialyl N-acetyllactosamine (Mochalova et al., 2007; Shtyrya et al., 2009). Most NAs used for these substrate specificity studies were either recombinant enzymes or expressed by purified viruses.
Here, we report on a substrate specificity study of NAs on thirty-seven human and avian influenza A viruses using gradient purified and/or non-purified viral particles conducted by employing a high-throughput screening format using a library of twenty α2–3- or α2–6-linked para-nitrophenol-tagged sialyl galactosides Siaα2–3/6GalβpNP synthesized by a highly efficient one-pot three-enzyme system (Cao et al., 2009).This proof-of-principle study demonstrates that the obtained information can be used to design novel neuraminidase inhibitors that target a select group of NAs on influenza A viruses.
Results
Viruses analysis
As shown in Table 1, thirty-seven viral strains were used in this study. Among these viral strains, twenty-two were avian-origin influenza A viruses isolated from free-flying wild birds or domestic captive birds in different locations of California and at various times (1996, 1998, 2006, 2005, 2007, 2008) with different hemagglutinin (belonging to group 1 hemagglutinin H1, H5, H6, H12 or group 2 hemagglutinin H3, H4, H10) and neuraminidase subtypes (belonging to group 1 neuraminidase N1, N5, N8 or group 2 neuraminidase N2, N6, N7, and N9); ten were avian H9N2 subtype influenza A viruses either isolated from domestic birds in Hong Kong in different years (1978, 1979, 1988, 1993, 1997, 1998, 1999) or isolated from duck in Hong Kong in 1979 and adapted to quail or chicken; one was a human H9N2 influenza A virus HK/2108/03 isolated in 2003; and four were human influenza A viruses belonging to H1N1, H3N1 and H3N2. Among human influenza A viruses, A/Memphis71 is a reassortant carrying the hemagglutinin of A/Memphis/1/71 (H3) and the neuraminidase of A/Bellamy/42 (N1). H9N2 avian influenza A viruses from birds with demonstrated adaptation to mammals (Wan et al., 2008; Xing et al., 2011) were classified into a separate group in order to test the effect of the adaptation and interspecies transmission of viruses from avian to mammalian species on the substrate specificity of neuraminidase.
Table 1.
Influenza A viruses used in this study.
| Viral strain | Type | Abbreviation | |
|---|---|---|---|
| Avian influenza A viruses | |||
| N1 | A/northern shoveler/California/HKWF569/2007 | H3N1 | sh/HKWF569/07 |
| A/northern shoveler/California/HKWF216/2007 | H6N1 | sh/HKWF216/07 | |
| A/mallard/California/8212/2008 | H6N1 | mal/8212/08 | |
| A/mallard/ California/8322/2008 | H6N1 | mal/8322/08 | |
| A/green winged teal/California/8326/2008 | H1N1 | tel8326/08 | |
| N2 | A/northern shoveler/California/HKWF268/2007 | H6N2 | sh/HKWF268/07 |
| A/green winged teal/California/7972/2008 | H6N2 | tel/7972/08 | |
| A/mallard/California/8035/2008 | H5N2 | mal/8035/08 | |
| N5 | A/american wigeon/California/HKWF295/2007 | H6N5 | wi/HKWF295/07 |
| A/american wigeon/California/HKWF541/2007 | H6N5 | wi/HKWF541/07 | |
| A/american wigeon/California/8352/2008 | H12N5 | wi/8352/08 | |
| N6 | A/env/California/7896/2008 | H4N6 | dk/7896/08 |
| N7 | A/emu/California/2092/1996 | H10N7 | emu/2092/96 |
| A/northern shoveler/California/JN950/2006 | H10N7 | sh/JN950/06 | |
| A/greater white-fronted goose/California/HKWF446/2007 | H10N7 | gos/HKWF446/07 | |
| A/northern shoveler/California/HKWF848/2007 | H3N7 | sh/HKWF848/07 | |
| A/northern shoveler/California/HKWF1021/2007 | H3N7 | sh/HKWF1021/07 | |
| A/northern shoveler/California/HKWF1128/2007 | H2N7 | sh/HKWF1128/07 | |
| A/cinnamon teal/California/HKWF1111/2007 | H5N7 | tel/HKWF1111/07 | |
| A/mallard/California/6957/2008 | H10N7 | mal/6957/08 | |
| N8 | A/ring-necked duck/California/K90/2005 | H6N8 | dk/K90/05 |
| N9 | A/green winged teal/California/8204/2008 | H5N9 | tel/8204/08 |
| H9N2 avian | A/duck//Hong Kong/448/1978 | H9N2 | dk/448/78 |
| A/duck/Hong Kong/702/1979 | H9N2 | dk/702/79 | |
| A/quail/Hong Kong/A28945/1988 | H9N2 | Q/A28945/88 | |
| A/quail/Arkansas/29209-1/1993 | H9N2 | Q/29209-1/93 | |
| A/duck/Hong Kong/Y280/1997 | H9N2 | dk/Y280/97 | |
| A/chicken/Hong Kong/G9/1997 | H9N2 | ck/G9/97 | |
| A/pheasant/California/2373/1998 | pt/2373/98 | ||
| A/chicken/Hong Kong/SF3/1999 | H9N2 | ck/SF3/99 | |
| A/duck/Hong Kong/702/1979-quail adapted23 | H9N2 | QA23 | |
| A/duck/Hong Kong/702/1979-chicken adaptedA10 | H9N2 | Qa23CkA10 | |
| H9N2 humana | A/Hong Kong/2108/2003 (H9N2) | H9N2 | HK/2108/03 |
| Human influenza A virusesa | |||
| N1 | A/Puerto Rico/34/8 | H1N1 | A/PR8 |
| A/Memphis/71 | H3N1 | A/Mem71 | |
| N2 | A/Udorn/307/72 | H3N2 | A/Udorn72 |
| A/Philippines/2/82/X-79 | H3N2 | A/Philips | |
Human influenza A viruses are shown in italics.
Substrate specificity of neuraminidases from avian and H9N2 influenza viruses
Using twenty α2–3- and α2–6-linked sialyl galactosides including Siaα2–3/6GalβpNP containing N-acetylneuraminic acid (Neu5Ac), N-glycolylneuraminic acid (Neu5Gc), 2-keto-3-deoxy-D-glycero-D-galacto-nonulosonic acid (Kdn), or their C5-derivatives (Table 2) (Cao et al., 2009), a substrate specificity study of neuraminidases from different strains of viruses all normalized to the same HA-titer shows important similarities and differences (Table 3 and Fig. S1)
Table 2.
Structures of para-nitrophenol-tagged α2–3- and α2–6-linked sialyl galactosides Siaα2–3/6GalβpNP containing different sialic acid forms (Cao et al., 2009) used for substrate specificity studies of neuraminidases on the surface of viral particles.
| Number | Compounds | Structure |
|---|---|---|
| 1a–10a | Siaα2–3GalβpNP | 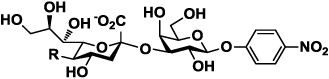 |
| 1b–10b | Siaα2–6GalβpNP | 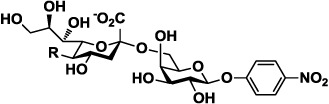 |
| Sialic acid name | Sialic acid structure | |
| 1a or 1b | Neu5Ac (R= NHAc) | 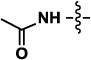 |
| 2a or 2b | Neu5AcF (R= NHCOCH2F) | 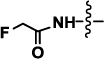 |
| 3a or 3b | Neu5AcOMe (R= NHCOCH2OMe) | 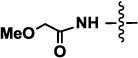 |
| 4a or 4b | Neu5AcN3 (R= NHCOCH2N3) |  |
| 5a or 5b | Neu5Gc (R= NHCOCH2OH) |  |
| 6a or 6b | 5d–Kdn (R= H) | |
| 7a or 7b | 5F–Kdn (R= F) | |
| 8a or 8b | 5Me-Kdn (R= OMe) | |
| 9a or 9b | 5N3-Kdn (R= N3) | |
| 10a or 10b | Kdn (R= OH) |
Table 3.
Substrate specificities of neuraminidases on avian and human influenza viruses.
| Type | Viral strain | Substrate Specificityb |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Comp. a | Comp.b | ||||||||
| a1a | 2a | 4a | 5a | 6a | 10a | 1b | 2b | ||
| N1 | sh/HKWF569/07 | 72 | ++++ | ++++ | ++++ | − | − | ++ | + |
| sh/HKWF216/07 | 64 | ++++ | +++ | ++++ | − | − | − | − | |
| mal/8212/08 | 60 | ++++ | +++ | ++++ | − | − | + | − | |
| mal/8322/08 | 52 | ++++ | ++ | +++ | − | − | − | − | |
| tel8326/08 | 88 | ++++ | + | ++++ | − | − | +++ | ++ | |
| N2 | sh/HKWF268/07 | 59 | ++++ | + | +++ | − | − | ++ | ++ |
| tel/7972/08 | 40 | +++ | − | +++ | − | − | ++ | + | |
| N5 | wi/HKWF295/07 | 41 | ++++ | ++ | +++ | − | − | + | − |
| wi/HKWF541/07 | 71 | ++++ | +++ | ++++ | − | + | ++ | + | |
| wi/8352/08 | 34 | ++++ | +++ | +++ | − | − | + | − | |
| N6 | dk/7896/08 | 28 | ++++ | + | ++ | − | − | ++ | + |
| N7 | emu/2092/96 | 32 | ++++ | − | +++ | + | + | ++ | + |
| sh/JN950/06 | 57 | ++++ | + | +++ | + | + | ++ | + | |
| gos/HKWF446/07 | 88 | ++++ | +++ | ++++ | ++ | +++ | +++ | +++ | |
| sh/HKWF848/07 | 88 | ++++ | +++ | ++++ | ++ | ++ | +++ | ++ | |
| sh/HKWF1021/07 | 78 | ++++ | +++ | ++++ | + | ++ | ++ | ++ | |
| sh/HKWF1128/07 | 46 | ++++ | ++ | +++ | + | + | ++ | + | |
| tel/HKWF1111/07 | 42 | ++++ | + | ++ | − | + | + | + | |
| mal/6957/08 | 81 | ++++ | + | ++++ | + | + | ++ | ++ | |
| N8 | dk/K90/05 | 72 | ++++ | + | +++ | − | − | − | − |
| H9N2 avian | dk/448/78 | 78 | ++++ | + | +++ | − | − | ++ | ++ |
| dk/702/79 | 76 | ++++ | ++ | ++++ | − | + | +++ | ++ | |
| Q/29209-1/93 | 84 | ++++ | ++ | ++++ | − | − | +++ | ++ | |
| pt/2373/98 | 73 | ++++ | − | +++ | − | − | ++ | + | |
| ck/SF3/99 | 69 | ++++ | − | +++ | − | − | ++ | ++ | |
| QA23 | 37 | ++++ | − | ++ | − | − | + | + | |
| Qa23CkA10 | 37 | ++++ | − | ++ | − | − | + | + | |
| H9N2 Humanc | HK/2108/03 | 32 | ++++ | − | +++ | − | − | +++ | ++ |
| Human unpurifiedc | |||||||||
| N1 | A/PR8 | 44 | ++++ | ++ | +++ | − | − | +++ | ++ |
| A/Mem71 | 35 | ++++ | ++ | +++ | − | − | +++ | ++ | |
| N2 | A/Udorn72 | 22 | ++++ | − | +++ | − | − | ++++ | +++ |
| A/Philips | 34 | ++++ | − | ++ | − | − | +++ | +++ | |
| Human purified N1c | A/PR8 (purified) | 84 | ++++ | ++ | ++++ | − | − | ++++ | +++ |
| A/Mem71(purified) | 82 | ++++ | +++ | ++++ | − | − | +++ | +++ | |
Absolute values for the percentage hydrolysis of Neu5Aca2–3GalppNP(compound 1a) and relative value for all other compounds. Relative values: Virus neuraminidase activity using Neu5Acα2–3GalβpNP (compound 1a) as the substrate is considered as 100%. "++++" represents activity ≥ 60%; "+++" represents activity ≥ 40% but < 60%; "++" represents activity ≥20% but <40%; "+" represents activity ≥10% but <20%; "-" represents activity < 10%.
The results for viruses with low activity [A405max is below 0.2 when Neu5Acα2–3GalβpNP (1a) is used as the substrate or the relative activity is below 10%]are not listed. These include viruses mal/8035/08 (H5N2), tel/8204/08 (H5N9), Q/A28945/88 (H9N2), dk/Y280/97 (H9N2), and ck/G9/97 (H9N2).
Human influenza A viruses are shown in italics. Underlined are group 1 NA viral strains. Others are group 2 NA viral strains.
All influenza A viruses tested have relatively good sialic acid cleavage activities towards α2– 3-linked sialosides Neu5Acα2–3GalβpNP (compound 1a), Neu5AcFα2–3GalβpNP (compound 2a), and Neu5Gcα2–3GalβpNP (compound 5a) although some viruses [A/mallard/California/8035/2008 (H5N2), A/green winged teal/California/8204/2008 (H5N9), A/Quail/Hong Kong/A28945/1988 (H9N2), A/Duck/Hong Kong/Y280/1997 (H9N2), A/Chicken/Hong Kong/G9/1997 (H9N2)] have weaker neuraminidase activities than others as shown by different absorption maxima indicated in the graphs (Fig. S1). All viruses tested have higher activity towards α2–3-linked sialosides (compounds 1a–10a) than α2–6-linked sialosides (compounds 1b-10b). Neu5Acα2–3GalβpNP (compound 1a) is the best substrate for all viruses tested. In contrast, α2–3- or α2–6-linked sialosides containing C5-OMe modified Neu5Ac (compounds 3a and 3b) and C5-F, -OMe, or –N3 modified Kdn (compounds 7a–9a, 7b–9b) are not effectively cleaved by any of the viral neuraminidases tested here. Among α2–6-linked sialosides tested (compounds 1b–10b), only the ones containing Neu5Ac or Neu5AcF (compound 1b and 2b) are suitable but weak substrates for the viral neuraminidases (Fig.S1).
Despite the similarities described above, the viruses showed different neuraminidase activities to the modifications on the C-5 hydroxyl group of sialic acid in the sialoside substrates. For example, α2–3-linked sialoside containing Neu5AcN3 (compound 4a), an azido-modified Neu5Ac, is a good substrate for some viral neuraminidases tested (N1, N5, most N7, and some N2) but not others (most N2, some N7, and N9). Many N7-expressing influenza A viruses cleaved α2–3-linked sialosides containing either Kdn (compound 10a) or its 5-deoxy derivative (5d–Kdn) (compound 6a), a unique substrate specificity not shared by the other strains tested.
H9N2 avian influenza A viruses continue to circulate worldwide. In Asia, H9N2 viruses have caused disease outbreaks and established lineages in land-based poultry (Aamir et al., 2007; Alexander, 2007; Guo et al., 1999; Kim et al., 2006; Naeem et al., 1999; Nili and Asasi, 2003). Importantly, H9N2 viruses have occasionally transmitted from land-based poultry to mammals, including humans. Mild respiratory diseases of humans were reported in Hong Kong and mainland China in 1999 and again in Hong Kong in 2003 (Butt et al., 2005; Lin et al., 2000; Peiris et al., 2001). We analyzed a series of H9N2 influenza A viruses isolated from ducks, quail, chicken, pheasant, and humans in Hong Kong over a period of 25 years (1978–2003) and two laboratory-adapted viruses (QA23 and Qa23CkA10) (Table 1). This provided an excellent opportunity to correlate possible changes of neuraminidase substrate specificity with interspecies transmission and avian to human adaptation (Wan et al., 2008). Similar to other N2 influenza A viruses (Fig. S1B), α2–3- and α2–6-linked sialosides containing Neu5Ac or Neu5AcF (compounds 1a, 1b, 2a, and 2b), and α2–3-linked sialoside containing Neu5Gc (compound 5a) were proven to be good substrates. However, the α2–3-linked sialoside containing Neu5AcN3 (compound 4a) was a weak substrate for some N2 strains such as dk/702/79 and Q/29209-1/93 (Fig. S1F) but not a substrate for others. No other compounds were substrates for these viruses. Significantly, there were noticeable differences between the overall sialidase activity and the relative ratio of α2–6-sialidase versus α2–3-sialidase activities among H9N2 strains isolated from different species and different times. The laboratory adapted H9N2 viruses showed lower relative α2–6-sialidase activity while the human H9N2 virus showed higher relative α2–6-sialidase activity (Fig. S1F).
Substrate specificities of neuraminidases from human influenza A viruses
A similar pattern of substrate specificity was observed for both sucrose-gradient-purified (Fig. S2A) and non-purified human influenza A viruses (Fig. S2B) (Table 3). The latter were used in form of virus-containing allantoic fluid. The purification process did, however, seem to decrease the neuraminidase activity of the purified viruses, requiring the use of higher viral amounts in the assays. Similar to the avian influenza A viruses tested, the neuraminidases from human viruses cleaved α2–3-linked sialosides with higher efficiency than α2–6-linked sialosides. All four human viruses cleaved Neu5Ac and Neu5AcF efficiently from α2–3- and α2–6-linked sialosides (compounds 1a, 1b, 2a, and 2b). α2–3-Linked sialoside containing Neu5Gc (compound 5a) is also a suitable substrate for these human virus strains. In contrast, α2–3-linked sialoside containing Neu5AcN3 (Neu5AcN3α2–3GalβpNP, compounds 4a) is a suitable substrate for A/PR8 and A/Mem71 (N1 type), but not for either A/Udorn72 or A/Philippines (N2 type). This result was consist with the NA activity of avian influenza A viruses described above. The human influenza A viruses tested were unable to cleave α2–3/6-linked sialosides containing Neu5AcOMe, Kdn, or Kdn derivatives (compounds 3a, 3b, 6a–10a, and 6b–10b). Overall, the sialic acid hydrolysis activities of human influenza A viruses were lower than those of avian influenza strains, as higher titers of human viruses (500–10,000 HAU mL−1) had to be used to achieve similar hydrolysis efficiency to avian viruses (12.8 HAU mL−1).
Inhibition studies of viral neuraminidases
A sialidase transition state analog, Neu5Ac2en, is a nonselective inhibitor against sialidases, including influenza A virus NAs. To test the inhibitory activity of the non-selective inhibitor Neu5Ac2en, we synthesized it and its C5-derivatives Neu5Ac2en derivatives 2-deoxy-2,3-dehydro-N-glycolylneuraminic acid (Neu5Gc2en), 2-deoxy-2,3-dehydro-N- azidoacetylneuraminic acid (Neu5AcN32en), and 2-deoxy-2,3-dehydro-Kdn (Kdn2en) (Li et al., 2011). These compounds were tested against NAs from different viruses, using a microtiter-plate based inhibition assay (Li et al., 2011) and their activities were compared to that of a commercially available influenza A virus NA inhibitor Zanamivir (a C4-guanadino derivative of Neu5Ac2en). As shown in Table 4, when Neu5Acα2–3GalβpNP (compound 1a) was used as the neuraminidase substrate for the inhibition assays, the IC50s of Neu5Ac2en were in the range of 2.6–48.3 µM for nine avian influenza A viruses with four NA subtypes and two human influenza A viruses with different NA subtypes. The inhibitory activity of Neu5Ac2en was the least efficient for avian influenza A viruses Q/29209-1/93 (IC50 = 48.3±5.8 µM) and mal/6957/08 (IC50 = 23.2±1.5 µM) and most efficient for avian influenza A viruses mal/8212/08 (IC50 = 3.8±0.4 µM) and wi/HKWF541/07 (IC50 = 1.8±0.5 µM), as well as human A/PR8 (IC50 = 2.6±0.5 µM). In comparison, commercial influenza virus specific inhibitor Zanamivir has IC50s in a range of 3.3–94.4 nM, indicating that Zanamivir is about three orders of magnitude more efficient than Neu5Ac2en. The Neu5Ac cleavage activities of Neu5Acα2–3GalβpNP by neuraminidases of human virus A/PR8 (IC50 = 3.3±0.2 nM) and avian viruses mal/8212/08 (IC50 = 6.5±0.3 nM) and Qa23CkA10 (IC50 = 9.5±1.6 nM) were more susceptible to Zanamivir inhibition than other viruses tested, especially avian viruses Q/29209-1/93 (IC50 = 59.9±3.6 nM) and mal/6957/08 (IC50 = 49.9±3.1 nM) as well as human virus A/Udorn72 (IC50 = 94.4±6.2 nM). When Neu5Acα2–6GalβpNP (compound 1b) was used as the neuraminidase substrate for some viruses which have significant α2–6-sialidase activity, the IC50s of Neu5Ac2en were in sub micromolar range (1.8–11.3 µM) and those of Zanamivir were in the range of 3.2–78.8 nM.
Table 4.
IC50s of Zanamivir and NeuAc2en for activities of human and influenza A virus neuraminidasesa
| Viral Strain | Type | IC50 |
|||
|---|---|---|---|---|---|
| Neu5Acα2–3GalβpNP | Neu5Acα2–6GalβpNP | ||||
| Zanamivir (nM) | Neu5Ac2en (uM) | Zanamivir (nM) | Neu5Ac2en (µM) | ||
| sh/HKWF569/07 | H3N1 | 14.3±1.6 | 10.7±1.4 | ND | ND |
| mal/8212/08 | H6N1 | 6.5±0.3 | 3.8±0.4 | ND | ND |
| wi/HKWF541/07 | H6N5 | 17.2±2.8 | 1.8±0.5 | ND | ND |
| sh/JN950/06 | H10N7 | 10.4±1.4 | 5.0±1.3 | ND | ND |
| gos/HKWF446/07 | H10N7 | 23.6±2.7 | 7.9±1.4 | ND | ND |
| mal/6957/08 | H10N7 | 49.9±3.1 | 23.2±1.5 | ND | ND |
| Q/29209-1/93 | H9N2 | 59.9±3.6 | 48.3±5.8 | 12.8±1.0 | 7.2±1.3 |
| ck/SF3/99 | H9N2 | 20.3±1.3 | 6.7±0.8 | ND | ND |
| Qa23CkA10 | H9N2 | 9.5±1.6 | 6.4±1.2 | ND | ND |
| A/PR8* | H1N1 | 3.3±0.2 | 2.6±0.5 | 3.2±0.5 | 1.8±0.4 |
| A/Udorn72* | H3N2 | 94.4±6.2 | 17.8±4.7 | 78.8±6.8 | 11.3±2.7 |
Unpurified avian influenza A viruses (12.8 HAU mL−1) and purified human viruses (A/PR8, 1 × 104 HAU mL−1; A/Udon, 5 × 103 HAU mL−1) were used. The reactions were carried out at 37 °C for 60 min with 1 mM of substrate.
Purified human influenza A virus; ND, not determined.
Because of the differences of virus NAs in their effectiveness of cleaving sialosides containing different C5-modified sialic acids, we hypothesized that C5-derivatives of the nonselective sialidase inhibitor Neu5Ac2en might act as selective inhibitors against NAs of certain influenza A strains. To test this hypothesis, the inhibitory activities of three C5-analogs of Neu5Ac2en: Neu5Gc2en, Neu5AcN32en, and Kdn2en, were tested against nine non-purified avian influenza A viruses and the two purified human influenza A viruses. As shown in Table 5, these three compounds were less potent inhibitors compared to Neu5Ac2en. Except for A/PR8 whose IC50 of Neu5Gc2en was around 100 µM, the IC50s of Neu5Gc2en for the other virus NAs were >100 µM. Furthermore, for all virus NAs tested, the IC50s of Neu5AcN32en were > 1 mM and those of Kdn2en were > 10 mM. The higher IC50 values obtained for Neu5Gc2en (IC50 > 100 µM), Neu5AcN32en (IC50 > 1 mM), and especially Kdn2en (IC50 > 10 mM) agreed well with the observation that sialosides containing Neu5Gc, Neu5AcN3, and Kdn were less efficient substrates than Neu5Ac-containing sialosides for influenza A viruses tested here (Fig. S1 and Fig. S2) .
Table 5.
Percentage inhibition of Neu5Gc2en, Neu5AcN32en, and Kdn2en against influenza A virus neuraminidases using Neu5Acα2–3GalβpNP or Neu5Acα2–6GalβpNP as the substrate.a
| Viral Strain | Type | % inhibition with Neu5Acα2–3GalβpNP as the substrate |
||||||
|---|---|---|---|---|---|---|---|---|
| Neu5Gc2en | Neu5AcN32en | Kdn2enb | ||||||
| 100 µM | 500 |µM | 1 mM | 1 mM | 5 mM | 10 mM | 10 mM | ||
| sh/HKWF569/07 | H3N1 | 24.1±5.6 | 56.3±0.7 | 72.9±0.3 | 27.6±0.3 | 51.9±0.1 | 61.6±1.3 | 7.5±0.4 |
| mal/8212/08 | H6N1 | 40.2±1.9 | 78.4±1.0 | 87.3±0.6 | 39.1±1.2 | 72.9±1.0 | 80.5±0.2 | 22.9±1.7 |
| wi/HKWF541/07 | H6N5 | 14.9±2.1 | 53.9±1.5 | 71.7±0.1 | 29.3±1.9 | 66.3±1.7 | 77.1±0.3 | 10.7±2.5 |
| sh/JN950/06 | H10N7 | 42.2±0.5 | 81.0±1.2 | 90.9±0.4 | 29.9±0.6 | 65.7±0.7 | 73.5±1.2 | 45.9±0.4 |
| gos/HKWF446/07 | H10N7 | 29.1±0.7 | 66.3±4.1 | 82.7±1.8 | 26.8±0.6 | 53.2±0.8 | 65.4±0.2 | 40.5±1.3 |
| mal/6957/08 | H10N7 | 14.3±2.0 | 51.6±1.5 | 71.9±0.2 | 8.6±0.4 | 34.2±0.4 | 52.2±2.0 | 18.8±0.9 |
| Q/29209-1/93 | H9N2 | 6.1±0.2 | 29.5±4.6 | 53.9±0.4 | 6.7±0.1 | 23.2±0.7 | 36.0±1.7 | – |
| ck/SF3/99 | H9N2 | 36.1±0.9 | 77.4±0.3 | 87.7±0.2 | 30.1±0.6 | 67.5±1.1 | 79.0±0.2 | 13.1±0.1 |
| Qa23CkA10 | H9N2 | 37.6±0.6 | 76.4±0.9 | 85.9±0.3 | 23.3±0.9 | 63.0±0.1 | 72.1±1.6 | – |
| A/PR8* | H1N1 | 48.5±2.2 | 84.6±0.2 | 90.4±0.2 | 66.7±0.5 | 88.9±0.9 | 90.1±0.4 | 42.2±2.4 |
| A/Udorn72* | H3N2 | 18.1±0.2 | 42.3±0.3 | 57.6±2.0 | – | – | 10.0±0.6 | – |
|
% inhibition with Neu5Acoα2–6Galβ p NP as the substrate |
||||||||
| Q/29209-1/93 | H9N2 | 39.4±0.5 | 80.2±1.27 | 90.5±0.4 | 23.8±2.2 | 57.3±0.3 | 64.2±1.4 | – |
| A/PR8* | H1N1 | 54.5±1.3 | 80.9±0.8 | 86.6±0.5 | 66.9±3.0 | 82.2±1.1 | 82.0±0.3 | 43.5±0.5 |
| A/Udorn72* | H3N2 | 24.4±0.3 | 61.7±0.3 | 73.6±0.3 | – | 24.4±0.8 | 43.5±2.4 | – |
Unpurified avian influenza A viruses (12.8 HAU mL−1) and purified human viruses (A/PR8, 1 × 104 HAU mL−1; A/Udorn72, 5 × 103 HAU mL−1; A/Mem71, 5 × 103 HAU mL−1; A/Philips: 2 × 104 HAU mL−1) were used. The reactions were carried out at 37°C for 60 min with 1 mM of substrate.
Kdn2en did not show any inhibition against the neuraminidase activity of A/Mem71 and A/Philips.
Purified human influenza A virus.
no inhibition
Discussion
Avian influenza A virus NAs have relatively high activity and exhibit substrate preference for α2–3-sialylated glycan substrates, while human virus NAs have lower activity and cleave both α2–3- and α2–6-sialylated glycans. Preferences between avian and human NAs differ also for the internal glycan structures of the sialoside substrates (Shtyrya et al., 2009). The binding of HAs (i.e. ligand specificity) to host cells needs to match the activity of NAs (i.e. substrate specificity) to achieve efficient viral infection and replication (Mitnaul et al., 2000; Shtyrya et al., 2009; Wagner et al., 2000). Despite the existence of more than 50 different natural sialic acid forms (Angata and Varki, 2002; Chen and Varki, 2010; Schauer, 2000), little information exists about the ligand preferences of HAs, or the substrate specificities of NAs on human and avian influenza A viruses with regard to the terminal sialic acid structures of sialosides.
Some similarities were identified among the NAs of all tested avian and human influenza A viruses. First, α2–3-linked sialosides were better substrates than α2–6-linked sialosides. Second, among 20 sialyl galactosides tested, Neu5Acα2–3GalβpNP (compound 1a) containing the most abundant sialic acid form, Neu5Ac, was the best substrate for all virus NAs. Third, substitution of one of the three hydrogen atoms by a fluorine atom on the N-acetyl group of Neu5Ac-terminated sialosides did not significantly decrease the activities of virus NAs, indicating the similarity between a hydrogen and fluorine. This property is very similar to that observed for human and bacterial sialidases (Cao et al., 2009; Li et al., 2011). Fourth, α2–3-linked sialoside containing a non-human sialic acid form, Neu5Gc, was a suitable substrate for all human and avian influenza A virus NAs tested, despite the lack of such monosaccharide at the site of host infection.
Despite these similarities, obvious differences existed between the degrees of substrate preference between the tested viral NAs for α2–6-linked sialosides versus α2–3-linked sialosides. Furthermore, N7 subtype NAs had distinct subtype specificities. They were the only viral NAs that used α2–3-linked sialoside substrates containing Kdn (compound 6a) or 5-deoxy-Kdn (5d–Kdn, compound 10a).
Our detailed analysis of the substrate specificity of 27 different influenza A virus NAs revealed that their relative preference for α2–3-linked sialoside substrates differed between host origin and time of isolation (Table S1). Although the ratios of relative neuraminidase activities on Neu5Acα2–6GalβpNP (compound 1b) versus Neu5Acα2–3GalβpNP (compound 1a) substrates were all below 1, they varied from low, such as for avian influenza A viruses sh/HKWF216/07 (H6N1, 0.09), mal/8322/08 (H6N1, 0.09), and dk/K90/05 (H6N8, 0.06), to high, as for human influenza A virus A/Udorn72 (H3N2, 0.64) and three other human viruses (1b/1a ratio > 0.4). In contrast to human influenza viral NAs, the ratios of activities on Neu5Acα2–6GalβpNP (compound 1b) versus Neu5Acα2–3GalβpNP (compound 1a) substrates were relatively low for most NAs of avian influenza A viruses except for gos/HKWF446/07 (H10N7, 0.57), dk/702/79 (H9N2, 0.43), Q/29209–1/93 (H9N2, 0.49), ck/G9/97 (H9N2, 0.48). These results were consistent with previous studies demonstrating that Neu5Acα2–3Gal was a preferred substrate over Neu5Acα2–6Gal for avian, swine, and N2 human influenza A viruses (Kobasa et al., 1999).
Based on their three-dimensional structures, neuraminidases from nine NA subtypes can be separated into two groups: Group 1 NA viral strains containing N1, N4, N5, and N8 subtypes and group 2 NA strains containing N2, N3, N6, N7, and N9 subtypes (Russell et al., 2006a). The major structural difference between group 1 and group 2 is a cavity observed in the active site close to ligand binding site of group 1 NAs, which is absent in the group 2 NAs (Russell et al., 2006a). Therefore, substrates for group 1 NAs might not be hydrolyzed by group 2 NAs. Based on their structural similarities, NAs in group 1 might have similar substrate specificities, and differ from those shared by group 2 NAs. Indeed, the substrate specificities of group 1 and 2 NAs revealed distinct patterns. As shown in Table 3, group 1 NAs have good cleavage activity against α2–3-linked sialosides containing Neu5AcF (compound 2a), Neu5AcN3 (compound 4a), and Neu5Gc (compound 5a), as well as Neu5Acα2–3GalβpNP (compound 1a). In contrast, group 2 NAs have good activity towards α2 –3-linked sialosides containing Neu5AcF (compound 2a), and Neu5Gc (compound 5a) as well as Neu5Ac α2–3GalβpNP (compound 1a). However, Neu5AcN3α2–3GalβpNP (compound 4a) is not a good substrate for most group 2 NAs, except for most N7 subtype strains and some belonging to the H9N2 subtype.
We demonstrate here that using a high-throughput approach to screen a library of para-nitrophenol-tagged α2–3- and α2–6-linked sialosides Siaα2–3/6GalβpNP is a highly effective means to conduct substrate specificity studies of sialidases from many influenza viruses. The neuraminidases of non-purified influenza A viruses share the same substrate specificity pattern as those from purified viral particles. Therefore, non-purified influenza A viruses can be directly used for substrate specificity assays, saving on labor and time-consuming viral isolation procedures and preventing the activity loss of NAs observed during the viral particle purification process. In addition, the inhibitory activity of two know NA inhibitors, Zanamivir and Neu5Ac2en, and three new candidate inhibitors, Neu5Gc2en, Neu5AcN32en and Kdn2en, were efficiently characterized using Neu5Acα2–3/6GalβpNP as substrate for viral NAs. These high-throughput substrate specificity and inhibition assays are much more efficient than the reported assays using peroxidase-linked lectin or fluorescence detection coupled with separation by an anion-exchange microcartridge (Mochalova et al., 2005) or a DEAE filter (Mochalova et al., 2007). The knowledge gained about the substrate specificity of individual influenza strains utilizing these rapid assays allows for the design of inhibitors that selectively target a distinct group of influenza A virus NAs.
Material and methods
Viruses
Thirty-seven avian and human influenza A virus strains were used in this study (Table 1). Unless indicated otherwise, the avian viruses were provided by the Cardona group and were collected through surveillance of wild birds done as part of Centers of Excellence in Influenza Research and Surveillance or Avian Influenza Coordinated Agricultural projects (A/northern shoveler/California/JN950/2006). A/emu/California/2092/1996 and A/pheasant/California/2373/1998 were from diagnostic cases submitted to the California Animal Health and Food Safety Laboratory. A human H9N2 virus preparation (A/Hong Kong/2108/2003) was provided by Dr. Yi Guan at the University of Hong Kong. All avian H9N2 subtype viruses excluding A/pheasand/California/2373/1998 were from Dr. Daniel Perez. Among H9N2 avian influenza A viruses, QA23 and Qa23CkA10 viruses are laboratory-adapted viruses. QA23 virus was developed by adapting a wild type duck H9N2 viral isolate, influenza dk/702/79 virus, in quail through 23 serial lung passages, and the chicken-adapted QA23CkA10 virus was obtained from the 10th chicken lung passage of the QA23 virus (Hossain et al., 2008). Allantoic-fluid containing the virus strains were used for the assays.
Among the strains used, four were human influenza A viruses (A/Puerto Rico/34/8, H1N1; A/Memphis71, H3N1; A/Udorn/307/72, H3N2; and A/Philippines/2/82/X-79, H3N2). A/Memphis71 is a reassortant influenza A virus strain carrying the hemagglutinin of A/Memphis/1/71 (H3) and the neuraminidase of A/Bellamy/42 (N1). All four human influenza A viruses used were propagated from frozen stocks originally obtained from Lorena Brown (University of Melbourne, Australia) and Charles Stephensen (UC Davis). Both sucrose-gradient purified virus particles and allantoic-fluid containing the human virus strains were used for the assays to test the effect of potential contaminating proteins in the allantoic fluid and/or the effects of the virus purification procedures on the activity and substrate specificity of viruses.
Virus propagation
Avian virus isolates were passaged by inoculation into 9–11 day specific-pathogen-free (SPF) embryonating chicken eggs (SPAFAS, Charles River) following standard methods (WHO, 2002). Allantoic fluid samples were tested for hemagglutination using 0.5% chicken blood (Colorado Serum Company, Denver, CO) as described previously (Li et al., 2008; WHO, 2002).
For human virus, fertilized hen eggs were incubated for 10 days with constant rotation at 37°C and 65% humidity. Eggs containing live embryos were infected with predetermined optimal concentrations of influenza virus strains by inoculation into the allantoic cavity and incubated for 2 days at 35°C followed by overnight incubation at 4°C. Allantoic fluid was harvested, batched, and centrifuged (8000 × g, 15 min at 4°C). Supernatants were aliquoted, snap-frozen on dry ice and stored at −80°C until used.
Purification of human influenza A virus
For virus purification, batches of allantoic fluid from infected eggs were centrifuged. Virus particles were precipitated from supernatants by overnight incubation at 4°C with 8% polyethylene glycol 6000 solution (Sigma- Aldrich, Dallas, TX) and pelleted by centrifugation (12,000 × g, 30 min at 4°C). To release the virus, the pellet was resuspended in phosphate buffered saline (PBS), sonicated, and centrifuged for 5 min at 3000 × g. The supernatant was collected and centrifuged (24,000 × g, 2 h at 4°C). The virus pellets were resuspended in a small volume of PBS and separated by sucrose gradient centrifugation (linear gradient 70–25%, 24,000 × g, 2 h at 4°C). The virus band was harvested, resuspended in PBS and centrifuged (24,000 × g, 2 h at 4°C). The pellet containing the virus was stored in a small volume of PBS at 4°C. The virus concentration (HAU/mL) was determined by standard hemagglutination assays using chicken red blood cells (Li et al., 2008; WHO, 2002).
Sialosides and neuraminidase inhibitors
The sialosides used in this study were synthesized using a one-pot three-enzyme approach as described previously (Cao et al., 2009). Zanamivir was from TCI America Ltd. (Portland, OR). Neuraminidase inhibitors 2-deoxy-2,3-dehydro-N-acetylneuraminic acid (Neu5Ac2en), 2-deoxy-2,3-dehydro-N-glycolylneuraminic acid (Neu5Gc2en), 2-deoxy-2,3-dehydro-N-azidoacetylneuraminic acid (Neu5AcN32en), and 2-deoxy-2,3-dehydro-Kdn (Kdn2en) were synthesized using a chemoenzymatic method as reported (Li et al., 2010).
Sialidase substrate specificity assays
Twenty sialyl galactosides including Siaα2–3/6GalβpNP containing N-acetylneuraminic acid (Neu5Ac), N-glycolylneuraminic acid (Neu5Gc), 2-keto-3-deoxy-D-glycero-D-galacto-nonulosonic acid (Kdn), or their C5-derivatives (Table 2) (Cao et al., 2009) were used to study the substrate specificity of neuraminidases on thirty-seven influenza A viruses using a microtiter plate-based high-throughput colorimetric assay method (Cao et al., 2009; Chokhawala et al., 2007). All sialidase assays were carried out at 37°C in duplicate in 384-well plates (Fisher Scientific, Chicago, IL) in a final volume of 20 µL each containing MES buffer (100 mM, pH 5.0), a certain titer of allantoic fluid containing virus or sucrose-purified viral particles, a sialoside substrate (0.3 mM), and an Aspergillus oryzae β-galactosidase (12 µg, 126 mU) from Sigma (St. Louis, MO). The amount of the β-galactosidase required to completely hydrolyze the GalβpNP within the time frame of the assay was pre-determined and confirmed by control assays with GalβpNP (0.3 mM). The reactions were carried out for 60 min and were stopped by adding pre-chilled CAPS buffer (N-cyclohexyl-3-aminopropane sulfonic acid, 40 µL, 0.5 M, pH 10.5). The amount of the para-nitrophenolate formed was determined by measuring the A405 nm of the reaction mixtures using a microtiter plate reader. As described before (Cao et al., 2009; Chokhawala et al. 2007), if a sialoside is the substrate of the neuraminidase of the virus tested, the sialic acid on the sialoside will be cleaved off by the neuraminidase to provide GalβpNP, which is quickly hydrolyzed by the excess amount of β-galactosidase in the reaction mixture to give para-nitrophenol. Addition of the pre-chilled CAPS buffer at the end of the incubation converts most of the para-nitrophenol to para-nitrophenolate whose concentration can be determined by measuring its absorption at 405 nm. The A405 nm reading is proportional to the amount of the sialic acid released from the sialoside by the neuraminidase of the virus and can be used to compare the substrate specificity of different neuraminidases on the surface of different viral strains.
Inhibition assay of viral neuraminidases
Inhibition assays were carried out in duplicate in 384-well plates in a total volume of 20 µL each in MES buffer (100 mM, pH 5.0) similar to that described above for the sialidase substrate specificity assays except that a fixed concentration of Neu5Acα2–3GalβpNP or Neu5Acα2– 6GalβpNP (1 mM) was used as the substrate and different concentrations of sialidase inhibitors were used. To determine IC50 values for Zanamivir and Neu5Ac2en, at least 11 different concentrations varying from 0 to 10.0 mM were used. IC50 values were obtained using Grafit 5.0 for logarithm concentration-response plots. When Neu5Gc2en (100 µM, 500 µM, and 1 mM), Neu5AcN32en (1 mM, 5 mM, and 10 mM), or Kdn2en (10 mM) was used as an inhibitor, only certain concentrations were chosen.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the provision of influenza A viruses for this work from Drs. Yi Guan, Daniel Perez and Peter Woolcock. This work was supported by NIH grant R01GM076360–04S1. X.C. is a Camille Dreyfus Teacher-Scholar and a UC-Davis Chancellor's Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aamir UB, Wernery U, Ilyushina N, Webster RG. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology. 2007;361(1):45–55. doi: 10.1016/j.virol.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25(30):5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 2002;102(2):439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Baum LG, Paulson JC. The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology. 1991;180(1):10–15. doi: 10.1016/0042-6822(91)90003-t. [DOI] [PubMed] [Google Scholar]
- Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 2005;43(11):5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Li Y, Lau K, Muthana S, Yu H, Cheng J, Chokhawala HA, Sugiarto G, Zhang L, Chen X. Sialidase substrate specificity studies using chemoenzymatically synthesized sialosides containing C5-modified sialic acids. Org. Biomol. Chem. 2009;7(24):5137–5145. doi: 10.1039/b916305k. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 2008;26(1):107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem. Biol. 2010;5(2):163–176. doi: 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokhawala HA, Yu H, Chen X. High-throughput substrate specificity studies of sialidases by using chemoenzymatically synthesized sialoside libraries. Chembiochem. 2007;8(2):194–201. doi: 10.1002/cbic.200600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couceiro JN, Baum LG. Characterization of the hemagglutinin receptor specificity and neuraminidase substrate specificity of clinical isolates of human influenza A viruses. Mem. Inst. Oswaldo Cruz. 1994;89(4):587–591. doi: 10.1590/s0074-02761994000400015. [DOI] [PubMed] [Google Scholar]
- Franca de Barros J, Jr, Sales Alviano D, da Silva MH, Dutra Wigg M, Sales Alviano C, Schauer R, dos Santos Silva Couceiro JN. Characterization of sialidase from an influenza A (H3N2) virus strain: kinetic parameters and substrate specificity. Intervirology. 2003;46(4):199–206. doi: 10.1159/000072428. [DOI] [PubMed] [Google Scholar]
- Guo Y, Li J, Cheng X. Discovery of men infected by avian influenza A (H9N2) virus. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1999;13(2):105–108. [PubMed] [Google Scholar]
- Hossain MJ, Hickman D, Perez DR. Evidence of expanded host range and mammalian-associated genetic changes in a duck H9N2 influenza virus following adaptation in quail and chickens. PLoS One. 2008;3(9):e3170. doi: 10.1371/journal.pone.0003170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinger D, Mochalova L, Chinarev A, Bovin N, Romanova J. Specificity of neuraminidase activity from influenza viruses isolated in different hosts tested with novel substrates. Arch. Virol. 2004;149(11):2131–2140. doi: 10.1007/s00705-004-0364-1. [DOI] [PubMed] [Google Scholar]
- Kim JA, Cho SH, Kim HS, Seo SH. H9N2 influenza viruses isolated from poultry in Korean live bird markets continuously evolve and cause the severe clinical signs in layers. Vet. Microbiol. 2006;118(3–4):169–176. doi: 10.1016/j.vetmic.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Kobasa D, Kodihalli S, Luo M, Castrucci MR, Donatelli I, Suzuki Y, Suzuki T, Kawaoka Y. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J. Virol. 1999;73(8):6743–6751. doi: 10.1128/jvi.73.8.6743-6751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Cardona CJ, Xing Z, Woolcock PR. Genetic and phenotypic characterization of a low-pathogenicity avian influenza H11N9 virus. Arch Virol. 2008;153(10):1899–1908. doi: 10.1007/s00705-008-0217-4. [DOI] [PubMed] [Google Scholar]
- Li Y, Cao H, Yu H, Chen Y, Lau K, Qu J, Thon V, Sugiarto G, Chen X. Identifying specific inhibitors against the human cytosolic sialidase NEU2 by substrate specificity studies. Mol. Biosyst. 2011 doi: 10.1039/c0mb00244e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. U. S. A. 2000;97(17):9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 2000;74(13):6015–6020. doi: 10.1128/jvi.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochalova L, Kurova V, Shtyrya Y, Korchagina E, Gambaryan A, Belyanchikov I, Bovin N. Oligosaccharide specificity of influenza H1N1 virus neuraminidases. Arch. Virol. 2007;152(11):2047–2057. doi: 10.1007/s00705-007-1024-z. [DOI] [PubMed] [Google Scholar]
- Mochalova LV, Korchagina EY, Kurova VS, Shtyria JA, Gambaryan AS, Bovin NV. Fluorescent assay for studying the substrate specificity of neuraminidase. Anal. Biochem. 2005;341(1):190–193. doi: 10.1016/j.ab.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Naeem K, Ullah A, Manvell RJ, Alexander DJ. Avian influenza A subtype H9N2 in poultry in Pakistan. Vet. Rec. 1999;145(19):560. doi: 10.1136/vr.145.19.560. [DOI] [PubMed] [Google Scholar]
- Nili H, Asasi K. Avian influenza (H9N2) outbreak in Iran. Avian Dis. 2003;47(3 Suppl):828–831. doi: 10.1637/0005-2086-47.s3.828. [DOI] [PubMed] [Google Scholar]
- Palese P, Tobita K, Ueda M, Compans RW. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61(2):397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary "human" H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 2001;75(20):9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006a;443(7107):45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- Russell RJ, Stevens DJ, Haire LF, Gamblin SJ, Skehel JJ. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj. J. 2006b;23(1–2):85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- Schauer R. Achievements and challenges of sialic acid research. Glycoconj. J. 2000;17(7–9):485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtyrya Y, Mochalova L, Voznova G, Rudneva I, Shilov A, Kaverin N, Bovin N. Adjustment of receptor-binding and neuraminidase substrate specificities in avian-human reassortant influenza viruses. Glycoconj. J. 2009;26(1):99–109. doi: 10.1007/s10719-008-9169-x. [DOI] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Spicuzza L, Spicuzza A, La Rosa M, Polosa R, Di Maria G. New and emerging infectious diseases. Allergy Asthma Proc. 2007;28(1):28–34. doi: 10.2500/aap.2007.28.2870. [DOI] [PubMed] [Google Scholar]
- Srinivasan A, Viswanathan K, Raman R, Chandrasekaran A, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Quantitative biochemical rationale for differences in transmissibility of 1918 pandemic influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 2008;105(8):2800–2805. doi: 10.1073/pnas.0711963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312(5772):404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Su B, Wurtzer S, Rameix-Welti MA, Dwyer D, van der Werf S, Naffakh N, Clavel F, Labrosse B. Enhancement of the influenza A hemagglutinin (HA)-mediated cell-cell fusion and virus entry by the viral neuraminidase (NA) PLoS One. 2009;4(12):e8495. doi: 10.1371/journal.pone.0008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne DE, Halvorson DA. In: Influenza in Diseases of poultry. 5th Ed. Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE, editors. Ames, IA: Blackwell Publishing; 2008. pp. 153–184. [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 2007;171(4):1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 2007;6(12):967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- Wagner R, Wolff T, Herwig A, Pleschka S, Klenk HD. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: a study by reverse genetics. J. Virol. 2000;74(14):6316–6323. doi: 10.1128/jvi.74.14.6316-6323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One. 2008;3(8):e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Response CDSa, editor. WHO. WHO manual on animal influenza diagnosis and surveillance. (5 ed.) 2002 WHO/CDC/CSR/NCS.
- Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Xing Z, Harper R, Anunciacion JD, Yang ZQ, Gao W, Qu B, Cardona CJ. Host immune and apoptotic responses to low pathogenic avian influenza virus H9N2 in human tracheobronchial epithelial cells. Am. J. Respir. Cell. Mol. Biol. 2011;44(1):24–33. doi: 10.1165/rcmb.2009-0120OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Newhouse EI, Amaro RE, Pao HC, Cheng LS, Markwick PR, McCammon JA, Li WW, Arzberger PW. Distinct glycan topology for avian and human sialopentasaccharide receptor analogues upon binding different hemagglutinins: a molecular dynamics perspective. J. Mol. Biol. 2009;387(2):465–491. doi: 10.1016/j.jmb.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


