Abstract
Purpose
To evaluate DNA synthesis and epigenetic modification in mouse oocytes during the first cell cycle following the injection of human or hamster sperm.
Methods
Mouse oocytes following the injection of human and hamster sperm and cultured in M16 medium.
Results
Male and female pronucleus formation, DNA synthesis, histone protein modification, and heterochromatin formation were similar in mouse oocytes injected with human or hamster sperm. However, DNA methylation patterns were altered in mouse oocytes following human sperm injection. Immunocytochemical staining with a histone H3-MeK9 antibody revealed that human and hamster sperm chromatin associated normally with female mouse chromatin, then entered into the metaphase and formed normal, two-cell stage embryos.
Conclusions
Although differences in epigenetic modification of DNA were observed, fertilization and cleavage occurred in a species non-specific manner in mouse oocytes.
Keywords: Interspecies ICSI, DNA synthesis, Epigenetic modification, Mouse oocyte
Introduction
The intracytoplasmic sperm injection (ICSI) technique is a powerful approach that can be used for studies in animal models to analyze fertilization processes and assisted human reproduction. Recently, injection of human sperm into mouse or rabbit oocytes has been used in studies relevant to assisted human reproduction [1, 2]. Successful two-cell division of mouse oocytes following the injection of human sperm has been observed [3]. However, it is not clearly understood whether cell cycle processes, epigenetic modification of male chromatin, metaphase entry, and cellular cleavage are maintained during interspecies fertilization by sperm injection.
During conventional in vitro fertilization, DNA replication is observed immediately after pronuclear formation, and it starts uniformly in both the male and female pronuclei in the intranuclear region and then in the peripheral regions of the nucleus and nucleolus [4]. In the mouse, single-cell embryos fertilized in vitro initiate DNA synthesis 6–8 h after insemination, and S phase lasts for 6–8 h [5]. In contrast, in human embryos DNA synthesis begins about 12 h after fertilization [6]. This difference in DNA synthesis onset may be due to cell cycle differences during mouse and human fertilization. In fact, two-cell cleavage is observed 14–15 h after ICSI in the mouse [7] and about 24 h after ICSI in human cells [8]. Although some information about pronuclear DNA replication is available for conventional mouse and human fertilization, little is known about the time of onset and end of replication in the mouse ICSI model following mouse or human sperm injection.
After sperm introduction into oocytes, the highly condensed sperm chromatin undergoes extensive nuclear remodeling and protamine-histone exchange, unlike nucleosomal maternal chromatin [9]. It is well known that several epigenetic modifications are involved in this reprogramming [10, 11]. The first observed epigenetic modifications of nucleosomal histones, including acetylation, methylation, phosphorylation and ubiquitination, are important chromatin alterations that broadly regulate gene transcription and silencing [12–14]. Modification can occur at several amino acid residues; a notable example is lysine 9 of histone H3 (H3K9), which can be acetylated or methylated [15]. In mouse zygotes, methylated H3K9 is distributed asymmetrically between the maternal and paternal pronuclei [16–20], but acetylated H3K9 is uniformly distributed in parental pronuclei shortly after fertilization [21, 22]. HP1, a heterochromatin protein 1 that interacts with methylated H3K9, is a component of silent chromatin at telomeres and the centrosome [23].
DNA methylation of genomic CpG dinucleotides is a major epigenetic modification, and it is associated with the repression of transcription and plays a key role in embryogenesis by silencing specific genes during development and differentiation. In mouse zygotes, the paternal genome undergoes active DNA demethylation shortly after fertilization, while maternal genomic DNA remains methylated throughout the first mitotic cycle [24, 25]. In contrast, paternal DNA demethylation cannot be detected in sheep, rabbit and goat zygotes [26–28]. Beaujean et al. [9] used interspecies sperm injection between mouse and sheep to show that sperm demethylation in the mouse is not dependent on the oocyte environment. Therefore, paternal demethylation appears to vary among species and the underlying mechanism is still unclear.
In most animals, the penetrating sperm introduces paternal centrosome as well as genetic material into the egg. The paternal centrosome organizes a functional microtubular aster called sperm aster in combination with the maternal centrosomal components [29]. Previous results showed that the sperm aster lead to union of male and female pronuclei and to the formation of spindle for the first cell division [30–33]. In contrast, mouse gametes do not follow the paternal patterns of centrosome inheritance during fertilization. In the mouse, maternal-derived microtubules appeared to move both male and female pronuclei into the center of the oocyte. Previously, normal fertilization can be achieved by injecting isolated sperm heads into oocytes in the mouse [34], cattle [35], rabbit [36], human [37], and pig [38]. This suggests that complete fertilization does not require all sperm components including the sperm centrosome.
To address whether DNA synthesis and epigenetic modification are influenced by maternal components in mouse oocytes following interspecific sperm injection, we compared pronuclear formation, DNA synthesis, the methylation and acetylation patterns of H3K9, HP1-β localization, DNA methylation, and microtubule formation in mouse oocytes following injection of mouse, human, or hamster sperm. To confirm the incorporation of mouse, hamster and human sperm chromatin into zygotes capable of entering the first mitotic metaphase and undergoing two-cell division, we also used immunocytochemical staining with the histone H3-MeK9 antibody, which differentially visualizes male and female chromatin.
Materials and methods
Preparation of mouse oocytes
Female B6D2F1 mice, 8–10 weeks of age, were induced to superovulate by i.p. injection of 5 IU Pregnant Mare Serum Gonadotrophin (PMSG, INTERVET) followed 48 h later by i.p. injection of 5 IU human Chorionic Gonadotrophin (hCG, INTERVET). Mature oocytes were collected from oviducts 14–15 h after hCG injection and freed from cumulus cells by a 3-min treatment with 0.1% bovine testicular hyaluronidase (Sigma, St. Louis, MO) in M2 (Sigma) medium. The cumulus-free oocytes were thoroughly rinsed and kept in M16 (Sigma) medium before ICSI for up to 3 h at 37°C.
Preparation of spermatozoa and ICSI procedure
Preparation of spermatozoa of mouse, hamster and human was followed by previous work [1, 9, 39]. Except that dense sperm masses from the cauda epididymides of the mouse and hamster were placed at the bottom of a 1.5-ml centrifuge tube containing 300 μl M2 medium. The supernatant containing actively motile spermatozoa was then transferred to 500 μl microtubes and stored in liquid nitrogen (LN2). In hamsters, acrosomes were disrupted by freeze-thawing spermatozoa without cryoprotection [1]. ICSI was carried out according to Kimura and Yanagimachi, using a piezoelectric micropipette actuator [40]. A single mouse spermatozoon was drawn tail first into an injection pipette, and the head was separated from the tail by applying a few piezo (PrimeTech; Ibaraki, Japan) pulses to the neck region. A straw (0.5 ml) of human spermatozoa was thawed in a 37°C water bath for 1 min. Thawed human semen were placed at the bottom of a 1.5-ml centrifuge tube containing 500 μl phosphate-buffered saline (PBS) and then frozen and sonicated for 5 min to isolate sperm heads, which were immediately injected into oocytes.
Activation
Following sperm injection, the injected oocytes were activated by 10 mM SrCl2 in Ca2+-free CZB [41] medium for 40 min and cultured in M16 medium.
Indirect immunofluorescence and scanning confocal microscopy
For histone modification detection, embryos at different developmental phases generated by ICSI were washed in PBS containing 0.1% polyvinyl alcohol (PBS-PVA), fixed for 20 min in 3.7% paraformaldehyde in PBS, and permeabilized with 0.2% Triton X-100 in PBS-PVA for 30 min at room temperature. The fixed embryos were incubated in a blocking solution (0.1 M glycine, 1% FBS, 0.01% Triton X-100, 1% powdered milk, 0.5% BSA, 0.02% sodium azide) for 1 h at 37°C and incubated with appropriate primary antibodies diluted in PBS-PVA at 37°C for 90 min. The antibodies and the dilutions used were as follows: mouse monoclonal anti-acetyl lysine 9 of histone H3 (H3-AceK9, Abcam 12179) antibody (1:100), rabbit anti-dimethyl lysine 9 in histone H3 (H3-MeK9, Cell Signaling 9,753 L) antibody (1:100), mouse anti-HP1-β (Chemicon, MAB3448) antibody (1:100), and mouse anti α-tubulin (Sigma) antibody (1:100).
For detection of DNA replication, embryos created by intracytoplasmic sperm head injection were cultivated in M16 containing 100 mM 5-bromo-2’-deoxyuridine (BrdU, Sigma; B9258) at 37°C in a humidified atmosphere of 5% CO2 in air. Embryos were then fixed with 3.7% paraformaldehyde and 0.5 N NaOH in PBS for 15 min at room temperature. They were next incubated for 1 h with a mouse anti-BrdU monoclonal immunoglobulin G (IgG) (Sigma; B2531) antibody diluted 50-fold with PBS.
After extensive washing in PBS-PVA containing 0.5% Triton X-100 and 0.5% BSA, the embryos were incubated in a blocking solution for 1 h at 37°C and labeled with a secondary FITC-conjugated (Invitrogen) antibody diluted 1:100 for 1 h at 37°C. The nuclear status of embryos was evaluated by staining with 1 μg/ml Hoechst 33,342 for 40 min. Following extensive washing, samples were mounted on slides.
For detection of DNA methylation, embryos were fixed in 3.7% paraformaldehyde in PBS for 30 min at 4°C, washed three times in PBS-PVA, and then permeabilized with 0.25% Triton X-100 in PBS-PVA for 1 h at room temperature. The embryos were treated with 2 N HCI for 40 min at room temperature and 100 mM Tris-HCI buffer (pH 8.0) [42], for 30 min at room temperature, and were then blocked in 5% BSA-PBS for 1 h. An anti-5-methylcytosine (Calbiochem; NA81) antibody (1:200) were incubated in the blocking solution for 2 h at 37°C, followed by several washes in PBS-PVA. After extensive washing, oocytes were stained with FITC-conjugated (Invitrogen) antibody (1:400) for 1 h at 37°C, followed by several washes. The nuclear status of embryos was evaluated with 10 μg/ml propidium iodide (PI, Sigma) for 1 h. Following extensive washing, the samples were mounted on slides.
Slides were examined using laser-scanning confocal microscopy, with a Leica DM IRB equipped with a krypton-argon ion laser for the simultaneous excitation of protein fluorescence and of PI or Hoechst 33,342 for DNA detection.
Statistical analysis
The general linear models procedure in the Statistical Analysis System [43] was used to analyze data from all experiments. Significant differences were determined using Tukey’s multiple range test [44] and P < 0.05 was considered significant.
Results
Male and female pronuclear formation was determined in mouse oocytes after 6 h following injection with mouse, hamster, or human sperm. Pronuclear formation and two cell divisions were observed for zygotes created with mouse, hamster, or human sperm, and more than 90% of the surviving injected eggs showed signs of normal fertilization. However, interspecies embryos were arrested at two cell stage (Table 1).
Table 1.
Development 2PN of mouse oocytes following mouse, hamster and human sperm injection
| Total no. Oocyte (r)a | No. of embryos developed | ||||
|---|---|---|---|---|---|
| 2PN (%) | 2-cell embryo (%) | 4-cell embryo (%) | Morula (%) | Blastocyst (%) | |
| 147 (3) | 145 (98.7 ± 2.31)b | 141 (96.0 ± 1.95)b | 136 (92.6 ± 4.98)b | 131 (89.3 ± 6.98)b | 111 (75.5 ± 5.08)b |
| 126 (3) | 120 (95.3 ± 2.21)b | 114 (90.5 ± 2.07)b | 2 (1.4 ± 3.47)c | 0 (0)c | 0 (0)c |
| 129 (3) | 123 (95.4 ± 2.20)b | 118 (91.6 ± 4.57)b | 6 (4.5 ± 4.55)c | 0 (0)c | 0 (0)c |
aNumber of replicates.
b,cValues with different superscripts in the same column differ from each other at P < 0.05
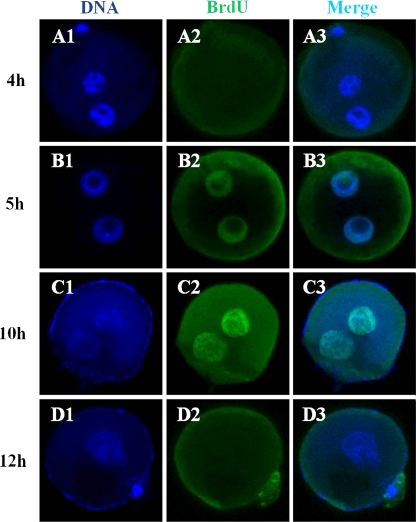
DNA replication after mouse sperm injection was evaluated by BrdU incorporation into pronuclei (Fig. 1). Replication was first observed in both male and female pronuclei 5 h after ICSI (Fig. 1B1-3). At this time, DNA synthesis was seen in peripheral areas of male and female pronuclei (Fig. 1B-2). From 6 to 10 h after ICSI, DNA synthesis was detected throughout both pronuclei (Fig. 1C1-3). At 12 h, incorporation of BrdU was not evident in either male or female pronuclei (Fig. 1D1-3). These observations were similar to those made for embryos created by injection with hamster or human sperm.
Fig. 1.
Localization of DNA replication sites in single-cell embryos created by injection of mouse oocytes with mouse sperm. Embryos were fixed at various time after ICSI (4, 5, 10 and 12 h) and stained with Hoechst 33342 (blue; A1, B1, C1 and D1) to detect nuclear DNA, and with an FITC-conjugated secondary antibody to detect anti-BrdU labeling (green; A2, B2, C2 and D2)
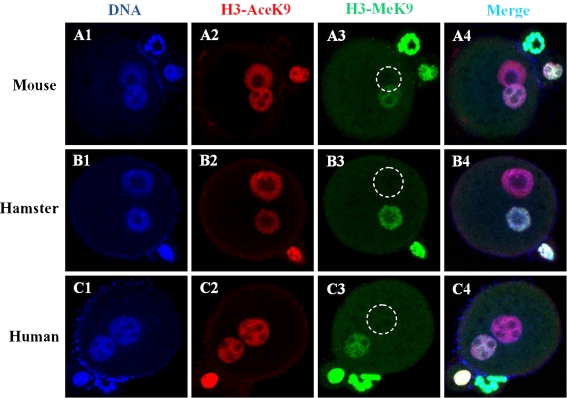
We investigated the H3K9 acetylation and methylation status of mouse, hamster and human sperm injected into mouse oocytes by immunocytochemistry (Fig. 2). When examined 6 h post-ICSI, reconstructed embryos produced by intra- and inter-specific ICSI procedures were similar, in that H3K9 was hyperacetylated in both parental pronuclei (Fig. 2A2, B2 and C2), but dimethylated H3K9 was asymmetrically distributed and, compared to the maternal pronucleus, the paternal pronucleus was not stained (Fig. 2A3, B3 and C3).
Fig. 2.
Reprogramming of H3K9 acetylation and methylation in embryos derived from mouse, hamster and human sperm injected into mouse oocytes. Embryos were fixed 6 h after injection and immunostained with antibodies against H3-AceK9- (red) and H3- MeK9-labeled (green) secondary antibodies. DNA was stained with Hoechst 33,342. Both parental pronuclei derived from mouse oocytes injected with mouse, hamster, or human sperm (A2, B2 and C2) were positively stained for H3-AceK9. The two parental pronuclei were asymmetrically stained for H3-MeK9; the paternal pronucleus (marked by a dashed white circle) showed no staining (A3, B3 and C3)
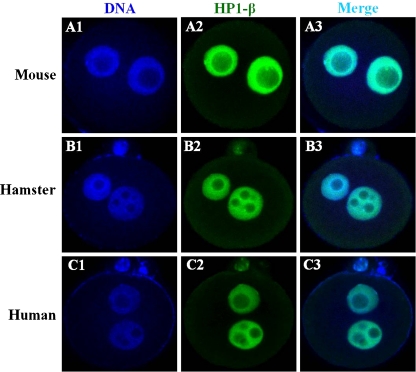
We then examined the localization of HP1-β produced in ICSI intra- and inter-specific zygotes (Fig. 3). As a reflection of chromatin organization, HP1-β was expressed 6 h after ICSI with mouse, hamster and human sperm (Fig. 3A2, B2 and C2).
Fig. 3.
Appearance and localization of HP1-β in zygotes derived from mouse, hamster and human sperm injected into mouse oocytes. Embryos were examined 6 h after ICSI by immunostaining with an HP1-β antibody (green; A2, B2 and C2). DNA was stained with Hoechst 33,342 (blue; A1, B1 and C1). HP1-β is detected in both pronuclei for mouse eggs after ICSI with mouse, hamster and human sperm
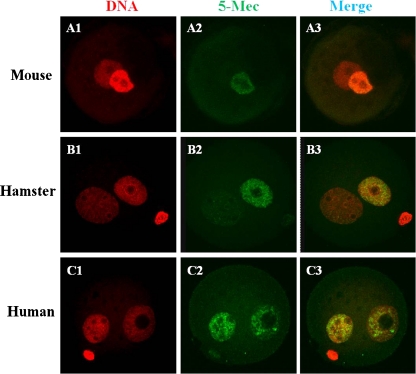
As shown in Fig. 4, the DNA methylation levels of the mouse and hamster paternal pronuclei in mouse oocytes were very similar, and 5-methyl-cytosine staining was not observed in paternal pronuclei 10 h after ICSI. However, DNA methylation persisted at least 10 h after mouse oocytes were injected with human sperm (Fig. 4C2).
Fig. 4.
Reprogramming of DNA methylation in zygotes derived from mouse, hamster and human sperm injected into mouse oocytes. Ten-hour embryos were stained with an anti-5-methyl-cytosine (5-MeC) antibody (green; A2, B2 and C2) antibody to detect DNA methylation and propidium iodide (PI) for nuclear DNA (red; A1, B1 and C1)
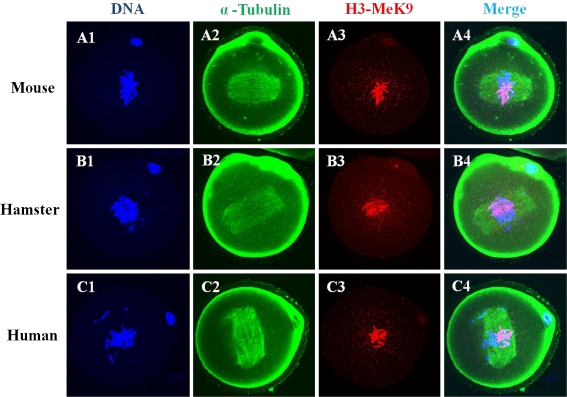
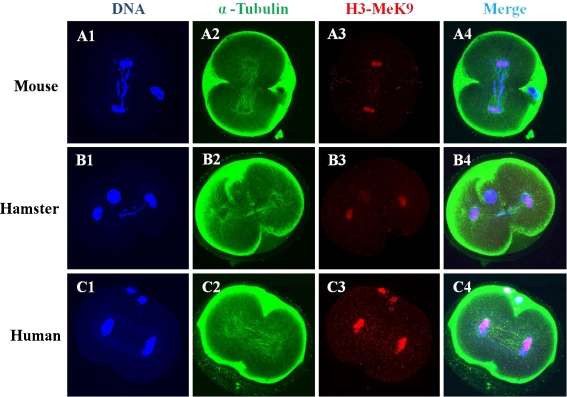
Microtubule organization and chromatin configuration in mouse zygotes after ICSI with mouse, hamster, or human sperm are shown in Figs. 5 and 6. Immunofluorescencestaining with the H3-MeK9 antibody allowed visualization of female chromatin. A normal mitotic metaphase and two cell divisions took place in mouse oocytes after ICSI with mouse, hamster, or human sperm.
Fig. 5.
Laser scanning confocal microscopic images of microtubules, H3K9 methylation and chromatin are shown at the metaphase stage of mouse zygotes after ICSI with mouse, hamster and human sperm. Metaphase chromatin from mouse oocytes after ICSI with mouse (A1-A4), hamster (B1-B4) and human (C1-C4) sperm was seen in half of the chromatin stained with H3-MeK9. All images are labeled for microtubules (green), DNA (blue) and H3-MeK9 (red)
Fig. 6.
Laser scanning confocal microscopic images of microtubules, H3K9 methylation and chromatin assembly at the telophase stage in mouse eggs after ICSI with mouse, hamster and human sperm. Telophase from mouse oocytes after ICSI with mouse (A1-A4), hamster (B1-B4) and human (C1-C4) sperm showed half of their chromatin stained with H3-meK9. All images are labeled for microtubules (green), DNA (blue) and H3- MeK9 (red)
Discussion
The microsurgical injection of a single spermatozoon into an oocyte, commonly called intracytoplasmic sperm injection or ICSI, has become the method of choice for the treatment of severe male factor infertility and the unexplained total failure of fertilization [45]. In the present study, we used a mouse model to analyze fertilization, DNA synthesis, epigenetic modification, metaphase entry and cell division in inter- and intra-specific embryos. The zygote which was got from ICSI may be different with the one in natural fertilization. It is difficult to compare the difference between the natural fertilization and ICSI, because the timing of natural fertilization is difficult to examine. Natural fertilization, which initiates a series of intracellular calcium ([Ca2+]) oscillations, releases oocytes from the MII arrest, and by reduced activity of maturation-promoting factor (MPF) and mitogen activation promoter factor (MAPK). Strontium also increased intracellular calcium concentrations; it caused multiple and periodic calcium oscillations. Efficient oocyte activation was obtained with strontium in oocytes from mice [46], pigs [47], and cows [48, 49]. Moreover, several study reported that oocytes activated using chemical substances or electroporation after ICSI [50–52]. Especially, in mice, oocytes injected with spermatozoa and treated with SrCl2 resulted in normal fertilization (92.9%) and good offspring rate (49.2%) of transferred embryos [50]. So we set the control group which injected with mouse sperm and then activated SrCl2.
To address whether pronuclei from interspecific sperm are functional in mouse oocytes and whether maternal DNA synthesis takes place, we monitored pronuclear formation and replication in mouse oocytes following injection with mouse, hamster, or human sperm. Previous studies of interspecific ICSI in pig [51, 52] and mouse oocytes found that male and female pronuclei successfully formed [53]. We found here that the incidence of male and female pronucleus formation was the same following the injection of mouse oocytes with mouse, human or hamster sperm. We also observed that DNA replication following mouse, human, or hamster sperm injection began in every instance at about 5 h and that normal male and female pronuclei formed 12 h after sperm injection. These observations suggest that the formation of functional pronuclei is not a species-specific process. Although the duration of the first cell cycle is about 15 h in the mouse and 24 h in human fertilization, DNA synthesis initiated and completed at the same time in mouse oocytes following mouse or human sperm injection. Our study suggests that DNA synthesis is mediated by maternal components.
In this study, we followed same methods of ICSI in mouse [9], hamster [1] and human [39]. We showed that H3K9 was hyperacetylated in both parental pronuclei (Fig. 2), but dimethylated H3K9 was asymmetrically distributed and, compared to the maternal pronucleus, the paternal pronucleus was not stained. In contrast, the presence of HP1-β was confirmed in all pronuclei with the anti-HP1-β antibody (Fig. 3). We also showed that inter-species ICSI oocytes displayed H3-AceK9, H3-MeK9 and HP1-β patterns similar to those of intra-species ICSI oocytes. Similar studies of mouse, sheep and human oocyte fertilization showed that paternal chromatin was associated with highly acetylated histones, as shown by anti-acetyl H3-AceK9 labeling, and that it was rapidly demethylated, as shown by anti-H3-MeK9 labeling [53, 54].
In general, DNA methylation is associated with gene silencing and heterochromatin formation. A few studies to compare the methylation status of embryos produced by intra- versus inter-specific ICSI have been conducted [9, 53, 55]. In this study, we observed that both pronuclei were relatively less demethylated after human sperm injection and that the male pronucleus was completely demethylated after mouse and hamster sperm injection. The result that the human-derived genome in mouse oocytes does not undergo active demethylation suggests that this process does not depend on maternal factors [56]. It was reported that mouse, bovine and sheep sperm show essentially the same demethylation trend in interspecific hybrids, in that paternal pronuclei are less demethylated in sheep and bovine oocytes, but with maximal or total demethylation in mouse oocytes [9]. The species differences might reflect variation in the efficiency of the demethylating factor, the presence or absence of activators/inhibitors, pronuclear size, or even distinct protein quantities of the putative demethylase [9]. Collectively, our results and those of previous studies suggest that sperm composition contributes to the reprogramming of DNA methylation.
In most animals, the paternal centrosome organizes a functional microtubular aster called sperm aster in combination with the maternal centrosomal components [52]. However, mouse and hamster gametes do not follow the paternal patterns of centrosome inheritance during fertilization. Furthermore, successful and normal fertilization can be achieved by injecting isolated sperm heads into mouse [34], cattle [35], rabbit [36], human [37], and pig [38] oocytes. This suggests that complete fertilization does not require all sperm components, including sperm centrosomes. Previously, Kim et al. [52] demonstrated incorporation of mouse sperm into metaphase entry in pig oocytes following ICSI. In this study, we demonstrated by staining H3K9 with the H3-MeK9 antibody, which stains only female chromatin, the incorporation of interspecific male chromatin into nuclei capable of metaphase entry and two-cell division in mouse oocytes.
In summary, we have observed fertilization processes in mouse oocytes following injection with mouse, hamster and human sperm. Although epigenetic modification of DNA differs in the paternal pronucleus, fertilization processes, including DNA synthesis and cleavage in mouse oocytes, occurred in a non-species specific manner. Our research results suggest that the mouse ICSI model using human sperm could be applied to determine the timing of events in human sperm during fertilization, which may have implications for managing assisted reproductive technology in the clinical context.
Acknowledgements
This study was supported financially by a grant from the BioGreen 21 program (201003010612240010400 and 201003010612240010500) and KOSEF (20100002070), Rural Development Administration (RDA), Republic of Korea.
Footnotes
Capsule Using mouse oocytes as ICSI recipients for human and mamster sperm, embryonic cell cycle progression is observed despite the presence of altered epigenetic modifications in sperm chromatin.
References
- 1.Yanagimachi R. Intracytoplasmic injection of spermatozoa and spermatogenic cells: its biology and application in human and animals. Reprod Biomed Online. 2005;10:247–88. doi: 10.1016/S1472-6483(10)60947-9. [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki T, Yamagata K, Baba T. Time-lapse and retrospective analysis of DNA methylation on mouse preimplantation embryos by live cell imaging. Dev Biol. 2007;304:409–19. doi: 10.1016/j.ydbio.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 3.Morozumi K, Yanagimachi R. Incorporation of the acrosome into the oocyte during intracytoplasmic sperm injection could be potentially hazardous to embryo development. Proc Natl Acad Sci USA. 2005;102:14209–14. doi: 10.1073/pnas.0507005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki E, Schultz RM. DNA replication in the 1-cell mouse embryo: stimulatory effect of histone acetylation. Zygote. 1999;7:165–72. doi: 10.1017/S0967199499000532. [DOI] [PubMed] [Google Scholar]
- 5.Akira K, Kwon OY, Tatsuo N, Tomohiro K. DNA synthesis in mouse 1-cell embryos containing transferred nuclei. J Reprod Dev. 1998;44:113–20. doi: 10.1262/jrd.44.113. [DOI] [Google Scholar]
- 6.Tesarík J, Kopecný V. Nucleic acid synthesis and development of human male pronucleus. J. Reprod Fert. 1989;86:549–58. doi: 10.1530/jrf.0.0860549. [DOI] [PubMed] [Google Scholar]
- 7.Probst AV, Santos F, Reik W, Almouzni G, Dean W. Structural differences in centromeric heterochromatin are spatially reconciled on fertilization in the mouse zygote. Chromosoma. 2007;116:403–15. doi: 10.1007/s00412-007-0106-8. [DOI] [PubMed] [Google Scholar]
- 8.Shoukir Y, Campana A, Farley T, Sakkas D. Early cleavage of in-vitro fertilized human embryos to the 2-cell stage: a novel indicator of embryo quality and viability. Hum Reprod. 1997;12:1531–6. doi: 10.1093/humrep/12.7.1531. [DOI] [PubMed] [Google Scholar]
- 9.Beaujean N, Taylor JE, McGarry M, Gardner JO, Wilmut I, Loi P, Ptak G, Galli C, Lazzari G, Brid A, Young LE, Meehan RR. The effect of interspecific oocytes on demethylation of sperm DNA. Proc. Natl. Acad. Sci USA. 2004;101:7636–40. doi: 10.1073/pnas.0400730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 11.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 12.Kouzarides T. chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 14.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–62. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 16.Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol. 2002;46:317–20. [PubMed] [Google Scholar]
- 17.Liu H, Kim JM, Aoki F. Regulation of histone H3 lysine 9 methylation in oocytes and early pre-implantation embryos. Development. 2004;131:2269–80. doi: 10.1242/dev.01116. [DOI] [PubMed] [Google Scholar]
- 18.Lepikhov K, Walter J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol. 2004;4:12. doi: 10.1186/1471-213X-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos S, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280:225–36. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Yeo S, Lee KK, Han YM, Kang YK. Methylation changes of lysine 9 of histone H3 during preimplantation mouse development. Mol cells. 2005;20:423–8. [PubMed] [Google Scholar]
- 21.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–25. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- 22.Worrad DM, Turner BM, Schultz RM. Temporally restricted spatial localization of acetylated isoforms of histone H4 and RNA polymerase II in the 2cell mouse embryo. Development. 1995;121:2949–59. doi: 10.1242/dev.121.9.2949. [DOI] [PubMed] [Google Scholar]
- 23.Yamazaki T. kobayakawa S, Yamagata K, Abe K, Baba T: Molecular dynamics of heterochromatin protein 1beta, HP1beta, during mouse preimplantation development. J Reprod Dev. 2007;53:1035–41. doi: 10.1262/jrd.19059. [DOI] [PubMed] [Google Scholar]
- 24.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 25.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–8. doi: 10.1016/S0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 26.Beaujean N, Hartshorne G, Cavilla J, Taylor J, Gardner J, Wilmut I, Meehan R, Young L. Non-conservation of mammalian preimplantation methylation dynamics. Curr Biol. 2004;14(7):R266–7. doi: 10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Shi W, Dirim F, Wolf E, Zakhartchenko V, Haaf T. Methylation reprogramming and chromosomal aneuploidy in in vivo fertilized and cloned rabbit preimplantation embryos. Biol Reprod. 2004;71:340–7. doi: 10.1095/biolreprod.103.024554. [DOI] [PubMed] [Google Scholar]
- 28.Hou J, Lei TH, Liu L, Cui XH, An XR, Chen YF. DNA methylation patterns in in vitro-fertilised goat zygotes. Reprod Fert Dev. 2005;17:809–13. doi: 10.1071/RD05075. [DOI] [PubMed] [Google Scholar]
- 29.Schatten G. The centrosome and its mode of inheritance: The reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- 30.Long CR, Pinto-Correia C, Duby RT. Ponce de Leon FA, Boland MP, Roche JF, Robl JM: Chromatin and microtubule morphology during the first cell cycle in bovine zygotes. Mol Reprod Dev. 1993;36:23–32. doi: 10.1002/mrd.1080360105. [DOI] [PubMed] [Google Scholar]
- 31.Navara CS, First NL, Schatten G. Microtubule organization in the cow during fertilization, polyspermy, parthenogenesis, and nuclear transfer: The role of the sperm aster. Dev Biol. 1994;162:29–40. doi: 10.1006/dbio.1994.1064. [DOI] [PubMed] [Google Scholar]
- 32.Kim N-H, Simerly C, Funahashi H, Schatten G, Day BN. Microtubule organization in porcine oocytes during fertilization and parthenogenesis. Biol Reprod. 1996;54:1397–404. doi: 10.1095/biolreprod54.6.1397. [DOI] [PubMed] [Google Scholar]
- 33.Kim N-H, Chung KS, Day BN. The role and distribution of microtubule and microfilaments during fertilization and parthenogenesis. J Reprod Fertil. 1997;111:143–9. doi: 10.1530/jrf.0.1110143. [DOI] [PubMed] [Google Scholar]
- 34.Kuretake S, Kimura Y, Hoshi K, Yanagimachi R. Fertilization and development of mouse oocytes injected with isolated sperm heads. Biol Reprod. 1996;55:789–95. doi: 10.1095/biolreprod55.4.789. [DOI] [PubMed] [Google Scholar]
- 35.Keefer CL. Fertilization by sperm injection in the rabbit. Gamete Res. 1989;22:59–69. doi: 10.1002/mrd.1120220107. [DOI] [PubMed] [Google Scholar]
- 36.Bourne H, Richings N, Liu DY, Clarke GN, Harari O, Baker HWG. Sperm preparation for intracytoplasmic injection: methods and relationship to fertilization results. Reprod Fert Dev. 1995;7:177–83. doi: 10.1071/RD9950177. [DOI] [PubMed] [Google Scholar]
- 37.Mansour RT, Aboulghar MA, Serour GI, Salah I. Intracytoplasmic injection of sperm head only. J Assist Reprod Genet. 1995;12(suppl):193. [Google Scholar]
- 38.Kim N-H, Lee JW, Jun SH, Lee HT, Chung KS. Fertilization of porcine oocytes following intracytoplasmic spermatozoon or isolated sperm head injection. Mol Reprod Dev. 1998;51:436–44. doi: 10.1002/(SICI)1098-2795(199812)51:4<436::AID-MRD11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura S, Terada Y, Horiuchi T, Emuta C, Murakami T, Yaegashi N, Okamura K. Human sperm aster formation and pronuclear decondensation in bovine eggs following intracytoplasmic sperm injection using a piezo-driven pipette: A novel assay for human sperm centrosomal function. Biol Reprod. 2001;65:1359–63. doi: 10.1095/biolreprod65.5.1359. [DOI] [PubMed] [Google Scholar]
- 40.Kimura Y, Yanagimachi R. Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development. 1995;121:2397–405. doi: 10.1242/dev.121.8.2397. [DOI] [PubMed] [Google Scholar]
- 41.Kishigami S, Wakayama S, Thuan NV, Ohta H, Mizutani E, Hikichi T, Bui HT, Balbach S, Ogura A, Boiani M, Wakayama Y. Production of cloned mice by somatic cell nuclear transfer. Nat Protoc. 2006;1:125–38. doi: 10.1038/nprot.2006.21. [DOI] [PubMed] [Google Scholar]
- 42.Dean W, Santos F, Stojkovic M. zakhartchenko V, Walter J, Wolf E, Reik W: Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98:13734–8. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.User’s Guide: statistics, version 5. Cary: SAS; 1985. [Google Scholar]
- 44.Steel RGD, Torrie JH. Principles and procedures of statistics. New York: McGraw Hill Nook Co; 1980. [Google Scholar]
- 45.Steirgehem AC, Nagy Z, Joris H, Liu J, Staessen C, Smitz J, Wisanto A, Devroey P. High fertilization and implantation rates after intracytoplasmic sperm injection. Hum Reprod. 1993;8:1061–6. doi: 10.1093/oxfordjournals.humrep.a138192. [DOI] [PubMed] [Google Scholar]
- 46.Bos-Mikichi A, Wood MJ, Candy CJ, Whittingham DG. Cytogenetical analysis and developmental potential of vitrified mouse oocytes. Biol Reprod. 1995;53:780–5. doi: 10.1095/biolreprod53.4.780. [DOI] [PubMed] [Google Scholar]
- 47.Che L, Lalonde A, Bordignon V. Chemical activation of parthenogenetic and nuclear transfer porcine oocytes using ionomycin and strontium chloride. Theriogenology. 2007;67:1297–304. doi: 10.1016/j.theriogenology.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Méo SC, Leal CLV, Garcia JM. Activation and early parthenogenesis of bovine oocytes treated with ethanol and strontium. Ann Reprod Sci. 2004;81:35–46. doi: 10.1016/j.anireprosci.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Méo SC, Yamazaki W, Leal CLV, Oliveira JA, Garcia JM. Use of strontium for bovine oocyte activation. Theriogenology. 2005;63:2089–102. doi: 10.1016/j.theriogenology.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Suganuma R, Walden CM, Butters TD, Platt FM, Dwek RA, Yanagimachi R, Spoel AC. Alkylated imino sugars, reversible male infertility-inducing agents, do not affect the genetic integrity of male mouse germ cells during short-term tre atment despite induction of sperm deformities. Biol Reprod. 2005;72:805–13. doi: 10.1095/biolreprod.104.036053. [DOI] [PubMed] [Google Scholar]
- 51.Kim NH, Jun SH, Do JT, Uhm SJ, Lee HT, Chung KS. Intracytoplasmic injection of porcine, bovine, mouse or human spermatozoon into porcine oocytes. Mol Reprod Dev. 1999;53:84–91. doi: 10.1002/(SICI)1098-2795(199905)53:1<84::AID-MRD10>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 52.Kim BK, Cheon SH, Lee YJ, Choi SH, Cui XS, Kim NH. Pronucleus formation, DNA synthesis and metaphase entry in porcine oocytes following intracytoplasmic injection of murine spermatozoa. Zygote. 2003;11:261–70. doi: 10.1017/S0967199403002314. [DOI] [PubMed] [Google Scholar]
- 53.Fulka H, Barnetova I, Mosko T, Fulka J., Jr Epigenetic analysis of human spermatozoa after their injection into ovulated mouse oocytes. Hum Reprod. 2008;23:627–34. doi: 10.1093/humrep/dem406. [DOI] [PubMed] [Google Scholar]
- 54.Hou J, Liu L, Zhang J, Cui XH, Yan FX, Guan H, Chen YF, An XR. Epigenetic modification of histone 3 at lysine 9 in sheep zygotes and its relationship with DNA methylation. BMC Dev Biol. 2008;8:60. doi: 10.1186/1471-213X-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaitseva I, Zaitsev S, Alenina N, Bader M, Krivokharchenko A. Dynamics of DNA-demethylation in early mouse and rat embryos developed in vivo and In vitro. Mol Reprod Dev. 2007;74:1255–61. doi: 10.1002/mrd.20704. [DOI] [PubMed] [Google Scholar]
- 56.Park JS, Jeong YS, Shin ST, Lee KK, Kang YK. Dynamic DNA methylation reprogramming: active demethylation and immediate remethylation in the male pronucleus of bovine zygotes. Dev Dyn. 2007;236:2523–33. doi: 10.1002/dvdy.21278. [DOI] [PubMed] [Google Scholar]