Abstract
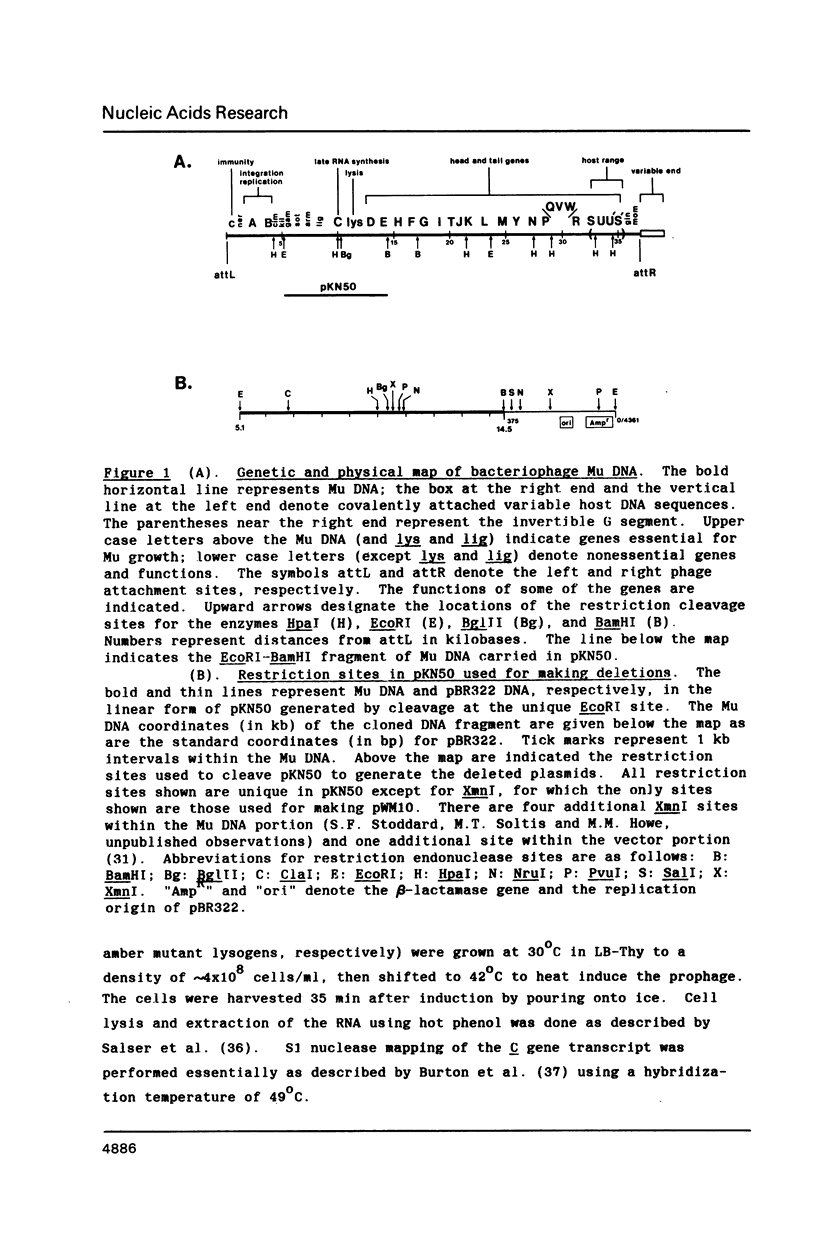
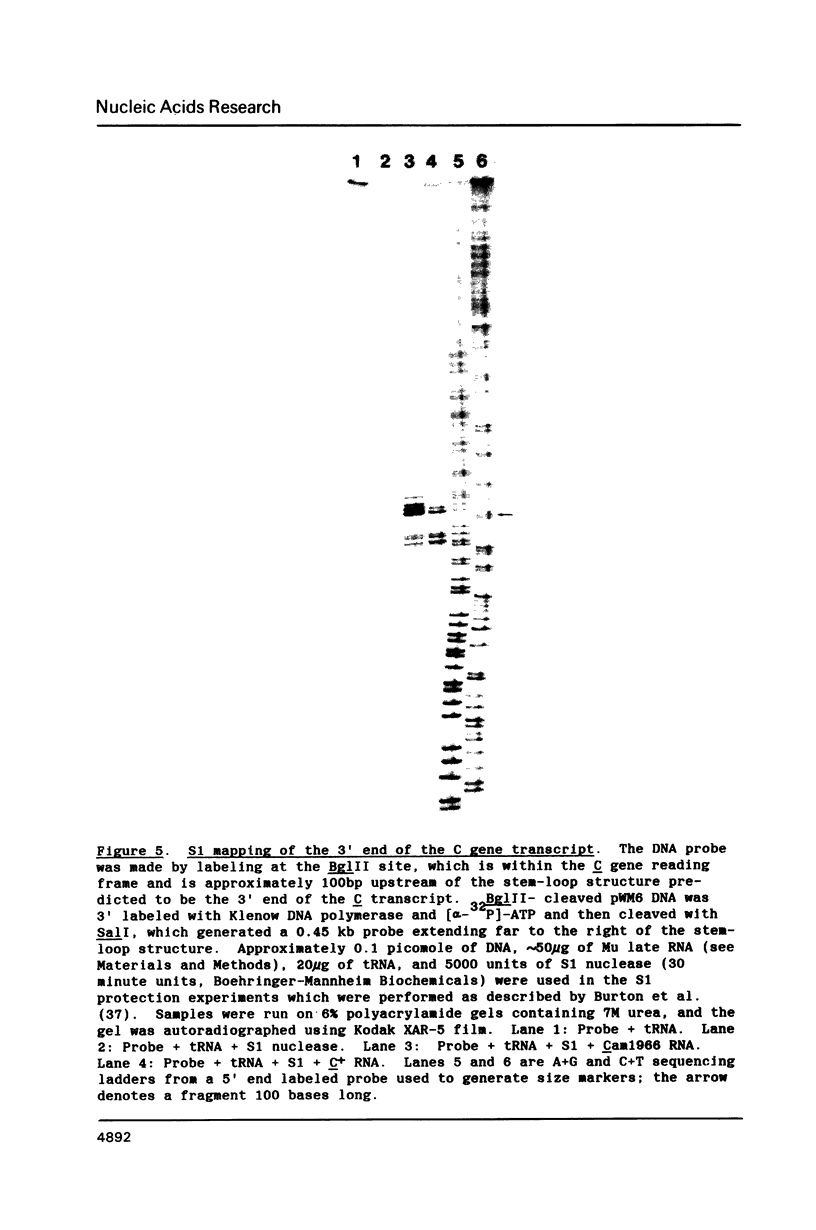
The C gene of bacteriophage Mu, required for transcription of the phage late genes, was localized by construction and analysis of a series of deleted derivatives of pKN50, a plasmid containing a 9.4 kb Mu DNA fragment which complements Mu C amber mutant phages for growth. One such deleted derivative, pWM10, containing only 0.5 kb of Mu DNA, complements C amber phages and transactivates the mom gene, one of the Mu late genes dependent on C for activation. The DNA sequence of the 0.5 kb fragment predicts a single long open reading frame coding for a 140 amino acid protein. Sequence analysis of DNA containing a C amber mutation located the base change to the second codon of this reading frame. Generation of a frameshift mutation by filling in a BglII site spanning codon 114 of this reading frame resulted in the loss of C complementation and transactivation activity. These results indicate that this open reading frame encodes the Mu C gene product. Comparison of the predicted amino acid sequence of the C protein with those of other transcriptional regulatory proteins revealed some similarity to a region highly conserved among bacterial sigma factors.
Full text
PDF














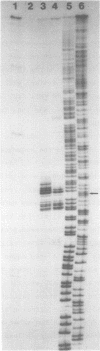
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Z. F., Gross C. A., Watanabe K. K., Burgess R. R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983 Feb;32(2):335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- Burton Z., Burgess R. R., Lin J., Moore D., Holder S., Gross C. A. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E coli K12. Nucleic Acids Res. 1981 Jun 25;9(12):2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMPBELL A. Sensitive mutants of bacteriophage lambda. Virology. 1961 May;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Brzustowicz L., Hannett N., Pero J. Bacteriophage SPO1 genes 33 and 34. Location and primary structure of genes encoding regulatory subunits of Bacillus subtilis RNA polymerase. J Mol Biol. 1984 Dec 15;180(3):533–547. doi: 10.1016/0022-2836(84)90026-3. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Pero J. Structure of a Bacillus subtilis bacteriophage SPO1 gene encoding RNA polymerase sigma factor. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1236–1240. doi: 10.1073/pnas.80.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelen M., Toussaint A. Isolation of conditional defective mutants of temperate phage Mu-1 and deletion mapping of the Mu-1 prophage. Virology. 1973 Jul;54(1):117–124. doi: 10.1016/0042-6822(73)90121-9. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Wijffelman C., Reeve J. Structural polypeptides and products of late genes of bacteriophage Mu: characterization and functional aspects. J Mol Biol. 1981 Jan 5;145(1):139–163. doi: 10.1016/0022-2836(81)90338-7. [DOI] [PubMed] [Google Scholar]
- Gitt M. A., Wang L. F., Doi R. H. A strong sequence homology exists between the major RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J Biol Chem. 1985 Jun 25;260(12):7178–7185. [PubMed] [Google Scholar]
- Gram H., Rüger W. Genes 55, alpha gt, 47 and 46 of bacteriophage T4: the genomic organization as deduced by sequence analysis. EMBO J. 1985 Jan;4(1):257–264. doi: 10.1002/j.1460-2075.1985.tb02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974 Sep 6;185(4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Howe M. M. Involvement of the invertible G segment in bacteriophage mu tail fiber biosynthesis. Virology. 1984 Apr 30;134(2):296–317. doi: 10.1016/0042-6822(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Howe M. M. Morphogenetic structures present in lysates of amber mutants of bacteriophage Mu. Virology. 1985 Jun;143(2):485–504. doi: 10.1016/0042-6822(85)90388-5. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J., Margolin W., Howe M. M. Regulation and expression of the bacteriophage mu mom gene: mapping of the transactivation (dad) function to the C region. Gene. 1985;39(1):71–76. doi: 10.1016/0378-1119(85)90109-x. [DOI] [PubMed] [Google Scholar]
- Hattman S. Specificity of the bacteriophage Mu mom+ -controlled DNA modification. J Virol. 1980 Apr;34(1):277–279. doi: 10.1128/jvi.34.1.277-279.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M., O'Day K. J., Schultz D. W. Isolation of mutations defining five new cistrons essential for development of bacteriophage Mu. Virology. 1979 Mar;93(2):303–319. doi: 10.1016/0042-6822(79)90235-6. [DOI] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Rothwell M. R., Higgins N. P. The early promoter of bacteriophage Mu: definition of the site of transcript initiation. Nucleic Acids Res. 1983 Aug 25;11(16):5483–5495. doi: 10.1093/nar/11.16.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R., Vaughn V., Lau E. T., VanBogelen R. A., Erickson J. W., Neidhardt F. C. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell. 1984 Aug;38(1):175–182. doi: 10.1016/0092-8674(84)90538-5. [DOI] [PubMed] [Google Scholar]
- Magazin M., Howe M., Allet B. Partial correlation of the genetic and physical maps of bacteriophage Mu. Virology. 1977 Apr;77(2):677–688. doi: 10.1016/0042-6822(77)90491-3. [DOI] [PubMed] [Google Scholar]
- Marrs C. F., Howe M. M. AvaII and BglI restriction maps of bacteriophage Mu. Virology. 1983 Apr 30;126(2):563–575. doi: 10.1016/s0042-6822(83)80013-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Merrick M. J., Gibbins J. R. The nucleotide sequence of the nitrogen-regulation gene ntrA of Klebsiella pneumoniae and comparison with conserved features in bacterial RNA polymerase sigma factors. Nucleic Acids Res. 1985 Nov 11;13(21):7607–7620. doi: 10.1093/nar/13.21.7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Day K., Schultz D., Ericsen W., Rawluk L., Howe M. Correction and refinement of the genetic map of bacteriophage Mu. Virology. 1979 Mar;93(2):320–328. doi: 10.1016/0042-6822(79)90236-8. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vrieling H., Van de Putte P. Transcription initiation of Mu mom depends on methylation of the promoter region and a phage-coded transactivator. Nature. 1983 Jan 27;301(5898):344–347. doi: 10.1038/301344a0. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Salser W., Gesteland R. F., Bolle A. In vitro synthesis of bacteriophage lysozyme. Nature. 1967 Aug 5;215(5101):588–591. doi: 10.1038/215588a0. [DOI] [PubMed] [Google Scholar]
- Schumann W., Bade E. G., Forgie R. A., Howe M. M. Cloning of DNA fragments of the right end of phage mu and location of the HindIII, SalI, PstI, and BamHI restriction sites on the genetic map of mu. Virology. 1980 Jul 30;104(2):418–425. doi: 10.1016/0042-6822(80)90344-x. [DOI] [PubMed] [Google Scholar]
- Schumm J. W., Moore D. D., Blattner F. R., Howe M. M. Correlation of the genetic and physical maps in the central region of the bacteriophage Mu genome. Virology. 1980 Aug;105(1):185–195. doi: 10.1016/0042-6822(80)90166-x. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Hoopes B. C., McClure W. R., Kleckner N. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983 Sep;34(2):673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- Somasekhar G., Szybalski W. Mapping of the Q-utilization site (qut) required for antitermination of late transcription in bacteriophage lambda. Gene. 1983 Dec;26(2-3):291–294. doi: 10.1016/0378-1119(83)90199-3. [DOI] [PubMed] [Google Scholar]
- Stragier P., Bouvier J., Bonamy C., Szulmajster J. A developmental gene product of Bacillus subtilis homologous to the sigma factor of Escherichia coli. Nature. 1984 Nov 22;312(5992):376–378. doi: 10.1038/312376a0. [DOI] [PubMed] [Google Scholar]
- Stragier P., Parsot C., Bouvier J. Two functional domains conserved in major and alternate bacterial sigma factors. FEBS Lett. 1985 Jul 22;187(1):11–15. doi: 10.1016/0014-5793(85)81203-5. [DOI] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Desmet L., Faelen M. Mapping of the modification function of temperate phage Mu-1. Mol Gen Genet. 1980 Jan;177(2):351–353. doi: 10.1007/BF00267450. [DOI] [PubMed] [Google Scholar]
- Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976 Mar;70(1):17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- Van Leerdam E., Karreman C., van de Putte P. Ner, a cro-like function of bacteriophage Mu. Virology. 1982 Nov;123(1):19–28. doi: 10.1016/0042-6822(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Lotterman B. Kinetics of Mu DNA synthesis. Mol Gen Genet. 1977 Mar 7;151(2):169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]
- Williams B. G., Blattner F. R., Jaskunas S. R., Nomura M. Insertion of DNA carrying ribosomal protein genes of Escherichia coli into Charon vector phages. J Biol Chem. 1977 Oct 25;252(20):7344–7354. [PubMed] [Google Scholar]
- van de Putte P., Cramer S., Giphart-Gassler M. Invertible DNA determines host specificity of bacteriophage mu. Nature. 1980 Jul 17;286(5770):218–222. doi: 10.1038/286218a0. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Giphart-Gassler M., Goosen N., Goosen T., van Leerdam E. Regulation of integration and replication functions of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):347–353. doi: 10.1101/sqb.1981.045.01.048. [DOI] [PubMed] [Google Scholar]