Abstract
In yeast, many genes are targeted to the nuclear periphery through interaction with the Nuclear Pore Complex upon activation. Targeting requires nucleoporin proteins and DNA elements in the promoters of these genes. We have recently found that targeting is regulated through the cell cycle. Immediately following the initiation of DNA replication, active genes lose peripheral localization for ∼30 minutes. This regulation is mediated by cyclic phosphorylation of a nucleoporin by Cdk1. Some genes that are targeted to the nuclear periphery upon activation remain at the nuclear periphery after repression, a phenomenon called transcriptional memory. Curiously, the mechanism that regulates localization of active genes to the nuclear periphery does not regulate the localization of the same genes after repression, suggesting that these genes are targeted by two distinct mechanisms. Finally, the localization of other parts of the genome at the nuclear periphery seems to be regulated by a distinct mechanism, suggesting that the spatial organization of the genome is coordinated with the progression of the cell cycle.
Key words: transcription, nucleus, nuclear pore complex, nuclear architecture, Cyclin dependent kinase, DNA replication
Introduction
The eukaryotic genome is spatially organized within the nucleus.1 Chromosomes in differentiated cells fold back on themselves and occupy well-defined territories within the nucleus.2 This organization is reproducible within cells of a particular tissue but is different between cells of different tissues. In many undifferentiated cells and in the brewers yeast Saccharomyces cerevisiae chromosomes assume a Rabl conformation, with telomeres associated with the nuclear envelope on one pole and centromeres associated with the nuclear envelope on the opposite pole.3 Global changes in gene positioning and chromosome folding occur during development, suggesting that the spatial organization of chromatin either impacts the regulation of gene expression or that it is a product of transcriptional status.
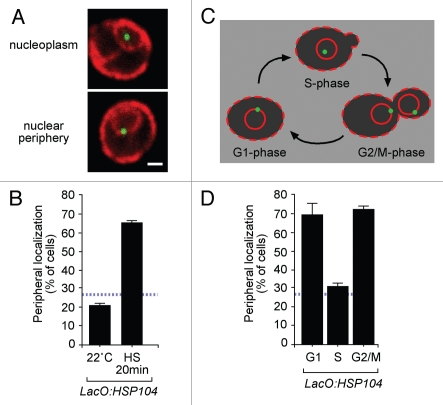
We are interested in determining the mechanisms that determine the localization of individual genes within the nucleus and how localization affects gene expression. In particular, we have focused on genes that relocalize from the nucleoplasm to the nuclear periphery upon activation.4–6 To address these questions, we have used a chromatin localization assay that allows us to determine the subnuclear localization of a particular gene with respect to the nuclear envelope.6,7 We integrate an array of Lac repressor binding sites beside the locus of interest in a strain expressing GFP-Lac repressor. We then perform immunofluorescence against GFP and a marker for the endoplasmic reticulum and nuclear envelope (Fig. 1A). We classify cells within the population as either nucleoplasmic or peripheral (Fig. 1A). A random distribution corresponds to ∼30% of the cells in the peripheral class (indicated as a blue hatched line in Fig. 1).6 After induction, many genes colocalize with the nuclear envelope in ∼65% of the population. For example, the HSP104 locus relocalizes from the nucleoplasm to the nuclear periphery upon heat shock (Fig. 1B).8
Figure 1.
Gene localization throughout the cell cycle. (A) Representative images of yeast cells labeled with GFP-Lac I (green) and Sec63-myc (red), classified as either nucleoplasmic or peripheral. Scale bar = 1 µm. (B) the localization of the HSP104 gene with respect to the nuclear periphery was quantified in populations of cells grown at 22°C or heat shocked at 37°C for 20 minutes. (C) Schematic representation of the staining of cells and gene positioning during the stages of the cell cycle: unbudded (G1), small-budded (S) and large-budded cells (G2/M). (D) the localization of HSP104 gene was quantified under activating conditions in unbudded, small-budded or large-budded cells from an asynchronous culture. In (B and D) the hatched line represents the level of colocalization of the lac repressor spot with the nuclear envelope predicted by chance.6
Several yeast genes localize at the nuclear periphery upon activation.4–6,9,10 The best characterized of these are the INO1 gene and the GAL1 gene.5,6 Localization at the nuclear periphery correlates with an interaction of the promoters of these genes with the nuclear pore complex (NPC)5,11 and requires nucleoporins.12–14 We have focused on the mechanism by which the INO1 gene is targeted to the NPC. We find that INO1 relocalizes to the nuclear periphery rapidly, independent of mRNA production12 and requires two DNA “zip codes” in the promoter called Gene Recruitment Sequences (GRSs15) and several components of the NPC (Nup1, Nup2, Nup60, Nup157, Nup42, Gle2) and associated proteins (Mlp2, Gcn5, Spt7, Spt20, Sus1, Sac3 and Thp1).15 Recruitment to the nuclear periphery promotes optimal levels of transcription of INO1 and HXK1 and may also couple gene expression with processing and export of mRNA molecules for translation in the cytoplasm.6,10,12,15
In our recent publication we have investigated the regulation of gene localization through the cell cycle.16 Since the yeast nucleus undergoes a closed mitosis with the nuclear membrane remaining intact through out the cell cycle, it is possible to analyze the association of a locus with the nuclear envelope throughout all of the stages of the cell cycle. Furthermore, because our marker for the nuclear envelope also marks the endoplasmic reticulum at the cortex of the cell (Fig. 1A and C), we can classify both nuclear localization and the bud morphology of each cell. In an asynchronous culture, the bud morphology can be used as an indication of cell cycle stage (Fig. 1C). Following mitosis, G1 cells are unbudded. Bud emergence occurs as DNA replication is initiated and S-phase cells have small buds measuring less than one-third the length of the mother cell's long axis. Cells with large buds (greater than one-third the length of the mother cell's long axis) are classified as G2/M phase (Fig. 1C). Using this refinement of the chromatin localization assay, we found that, in unbudded cells and large-budded cells, both the INO1 gene and the GAL1 gene localized at the nuclear periphery. In contrast, in the small-budded cells, INO1 and GAL1 localized in the nucleoplasm. We have observed this phenomenon for other genes that are targeted to the nuclear periphery. For example, when we examined the localization of HSP104 within the nucleus through the cell cycle, we found that it was regulated exactly as we had observed for INO1 and GAL1 (Fig. 1D). During G1 and G2/M, HSP104 localized at the nuclear periphery in 69% and 72% of the cells, respectively. However, in small-budded cells, HSP104 localized at the nuclear periphery in 31% of the cells (Fig. 1D). All genes we have examined to date that localize to the nuclear periphery when active have exhibited this behavior and cyclical association with the nuclear membrane through the cell cycle, indicating that this process may be a general property of gene recruitment (our unpublished results).
Consistent with the idea that INO1 and GAL1 lose peripheral targeting during S-phase, in cells arrested with hydoxyurea, both genes localized to nucleoplasm. This is in contrast to cells arrested during mitosis (either cdc20-1 or by treatment with nocodazole), in which INO1 and GAL1 remain at the nuclear periphery. Furthermore, in cdc6-1 mutant cells arrested at the restrictive temperature (37°C), both genes were found at the periphery, suggesting that the initiation of DNA replication was upstream of the loss of peripheral localization. After the cdc6-1 mutant was returned to the permissive temperature, INO1 and GAL1 relocalized to the nucleoplasm in less than 5 minutes. Thus, very quickly after the initiation of DNA replication, the genes lose peripheral localization.
The Nuclear Pore Complex
Previous work from our lab and others has clearly demonstrated a role for the nuclear pore complex in gene recruitment. We have created numerous deletion mutants for non-essential components of the NPC and found that several components of the nucleoplasmic basket are critical for INO1 recruitment to the nuclear periphery (e.g., Nup1, Nup2 and Nup60).15 Many of these components have been identified to be targets of the master regulator of the cell cycle, cyclin-dependent kinase (Cdk1, CDC28 in Saccharomyces cerevisiae). We hypothesized that phosphorylation of nuclear pore components by Cdk1 may regulate gene recruitment during the cell cycle. Indeed, using a temperature-sensitive mutant of Cdk1 (cdc28-1) we found that targeting of INO1 and GAL1 to the nuclear periphery required Cdk1 activity; both genes localized to the nucleoplasm in cells incubated at the restrictive temperature. This effect was independent of the phase of the cell cycle at which these cells were arrested; cdc28-1 cells arrested during mitosis with nocodazole and then shifted to the restrictive temperature showed the same loss of peripheral localization.
The nucleoporin Nup1, which is required for targeting of both INO1 and GAL1 to the nuclear periphery, is a Cdk1 target both in vivo and in vitro.17,18 We found two canonical Cdk1 phosphorylation sites (S/TPIK) at the amino terminus of Nup1. To determine if phosphorylation of these sites is required for targeting of INO1 and GAL1 we mutagenized the phosphoacceptor sites at S161 or T344 to cysteine residues. Although neither mutation alone had an effect on localization, the double mutant abolished recruitment to the periphery indicating that phosphorylation of Nup1 is required for gene targeting to the nuclear periphery. This double mutant was able to complement loss of Nup1 in other functional in other assays, suggesting that it was not a null mutant. Furthermore, substitution of aspartic acid residues for either phosphoacceptor residue resulted in loss of cell cycle regulation of gene localization. In cells expressing these mutant forms of Nup1, INO1 and GAL1 remained at the periphery during S phase. Finally, these mutant forms bypassed the requirement for Cdk1 in gene targeting, suggesting that Nup1 is the only substrate of Cdk1 important for this regulation.
Gene Positioning is a Form of Cellular Memory
In the course of our studies on the mechanism of gene recruitment to the nuclear periphery, we identified an unexpected consequence of gene positioning. We discovered that cells possess a form of memory that marks previously expressed genes and primes them for expression in the future.12 Some of the genes that are targeted to the nuclear periphery when expressed remain at the nuclear periphery for several generations after they are turned off. Surprisingly, the molecular mechanism that controls localization of INO1 at the nuclear periphery when it is active is different from the mechanism that controls localization of INO1 at the nuclear periphery after it is turned off. Retention after repression requires a different DNA zip code, different components of the nuclear pore complex and the histone variant H2A.Z.19 We have recently found that the interaction of a Memory Recruitment Sequence with the NPC after repression alters the chromatin structure of the promoter, allowing faster expression in the future.19 Therefore, transcriptional memory represents a novel regulatory mechanism by which previously expressed genes can be marked and primed for faster activation in the future. The proteins required for this phenomenon are universally conserved, raising the possibility that similar phenomena could occur in humans.
The localization of recently repressed INO1 and GAL1 at the nuclear periphery was not regulated during the cell cycle. In cells that have recently repressed INO1 or GAL1, the genes remain at the nuclear periphery throughout the cell cycle.16 Furthermore, loss of the critical Cdk1 phosphorylation sites on Nup1 had no effect on the localization of recently repressed INO1.16 This indicates that the molecular mechanism used for genes with transcriptional memory is unique from that used during active transcription.
Localization of Telomeres is Regulated by a Different Mechanism
Yeast telomeres are tethered to the nuclear envelope by a different mechanism.20–22 The localization of telomeres at the nuclear periphery promotes silencing of subtelomeric genes.23 The localization of telomeres at the nuclear periphery is also coordinated with the cell cycle although there are striking mechanistic differences. The tethering of yeast telomeres to the nuclear envelope is lost during G2, not S phase. Telomeres localize at the nuclear periphery in unbudded and small-budded cells and in the nucleoplasm in large-budded cells.20–22 Using several approaches to manipulate the length of S phase, Ebrahimi and Donaldson found that telomeres remained at the periphery during S phase but relocalized to the nucleoplasm as cells progressed towards mitosis.22 Similarly, the Gasser lab observed that telomeric clusters are maintained at the periphery when cells are arrested in S phase with hydroxyurea, but are lost as the cells enter mitosis 20 minutes after the drug is washed away.20 Loss of telomeric tethering during the cell cycle is mediated by loss of Sir4 and Ku binding to telomeres.22 Thus, the localization of different parts of the genome are independently regulated through the cell cycle, suggesting an elaborate coordination of the spatial organization of the genome with cyclic events such as DNA replication, chromosome condensation and chromosome segregation.
Conclusions
We have discovered a new phenomenon that is regulated through the cell cycle by Cdk1 phosphorylation of a single nuclear pore protein. The physiological significance of this regulation is not clear. Introduction of phosphomimetic mutations into Nup1 leads to loss of regulation and constitutive localization at the nuclear periphery throughout the cell cycle. However, these cells do not display dramatic phenotypes, indicating that, under typical lab conditions, this system is dispensable. However, we speculate that regulation of gene localization to the nuclear periphery may reduce physical conflict between DNA replication or repair and active transcription at the periphery. Future work will test this hypothesis.
Acknowledgements
The work described in this Extraview was supported by the Searle Leadership fund at Northwestern, National Institutes of Health grant GM080484 and a W.M. Keck Young Scholar in Medical Research Award.
Abbreviations
- Cdk
cyclin dependent kinase
- GRS
gene recruitment sequence
- HSP
heat shock protein
- NUP
nucleoporin
- MRS
memory recruitment sequence
References
- 1.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories: A functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: Positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 6.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brickner DG, Light WH, Brickner JH. Quantitative localization of chromosomal loci by immunofluorescence. In: Weissman J, Guthrie C, Fink GR, editors. Methods in Enzymology. Burlington: Academic Press; 2010. pp. 569–580. [DOI] [PubMed] [Google Scholar]
- 8.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, et al. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci USA. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 11.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: The nucleoporepromoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Brickner DG, Cajigas IC, Fondufe-Mittendorf Y, Ahmed S, Lee P-C, Widom J, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 14.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, et al. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed S, Brickner DG, Light WH, McDonough M, Froyshteter AB, Volpe T, et al. DNA zip codes control an ancient mechanism for targeting genes to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brickner DG, Brickner JH. Cdk phosphorylation of a nucleoporin controls gene localization through the cell cycle. Mol Biol Cell. 2010;21:3421–3432. doi: 10.1091/mbc.E10-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 18.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Light WH, Brand V, Brickner DG, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laroche T, Martin SG, Tsai-Pflugfelder M, Gasser SM. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J Struct Biol. 2000;129:159–174. doi: 10.1006/jsbi.2000.4240. [DOI] [PubMed] [Google Scholar]
- 21.Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- 22.Ebrahimi H, Donaldson AD. Release of yeast telomeres from the nuclear periphery is triggered by replication and maintained by suppression of Ku-mediated anchoring. Genes Dev. 2008;22:3363–3374. doi: 10.1101/gad.486208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–625. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]