Abstract
FoxM1 transcription factor (previously called HFH-11B, Trident, FoxM1b, Win and MPP2) is expressed in actively dividing cells and is critical for cell cycle progression. FoxM1 expression is induced in a variety of tissues during embryogenesis, and Foxm1-/- mice exhibit embryonic lethal phenotype due to multiple abnormalities in the liver, heart, lung and blood vessels. FoxM1 levels are dramatically decreased in adult tissues, but FoxM1 expression is re-activated during organ injury and numerous cancers. In this review, we discussed the role of FoxM1 in different cell lineages using recent data from transgenic mouse models with conditional “gain-of-function” and “loss-of-function” of FoxM1, as well as tissue samples from human patients. In addition, we provided experimental data showing additional sites of FoxM1 expression in the mouse embryo. Novel cell-autonomous roles of FoxM1 in embryonic development, organ injury and cancer formation in vivo were analyzed. Potential application of these findings for the diagnosis and treatment of human diseases were discussed.
Key words: winged helix DNA binding domain, forkhead transcription factor, FoxM1, embryonic development, cancer, cellular proliferation, cell cycle, transgenic mice
Introduction
The Forkhead Box (Fox) proteins belong to a large family of transcription factors that are evolutionary conserved in the Winged Helix/Forkhead DNA binding domain.1–3 The name of “Fork head” derives from two head-like structures in fork head mutant Drosophila embryos that exhibited defects in formation of anterior and posterior gut.4 Fox family includes more than 55 distinct mammalian members grouped into 17 subfamilies according to their sequence homology within the DNA binding domain.5 Several mutations in Fox genes have been linked to human congenital disorders. Mutations in FoxC1, FoxC2, FoxE3 and FoxL2 cause various eye abnormalities, whereas mutations in FoxE1 and FoxN1 are linked to thyroid hypoplasia, cleft palate and T-cell immunodeficiency.6 Heterozygous deletions or point mutations in FoxF1 gene locus were recently found in 30% of human patients with Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACD/MPV), a severe congenital disorder with mortality rate of 100% during the first months of life.7 FoxP2 mutations are associated with speech and language disorders, and FoxP3 mutations were found in patients with immune deficiency, enteropathy and complex endocrine abnormalities.6 Duplications of FoxG1 in 14q12 are associated with developmental epilepsy, mental retardation and severe speech impairment.8
Fox proteins play important roles in pathogenesis of human cancers. Chromosomal translocations of FoxO proteins have been identified as a causative factor in human alveolar rhabdomyosarcoma and acute lymphoid leukemia.9,10 The loss of FoxO1A through 13q14 chromosomal deletion was associated with prostate cancer and progression towards androgen independence.11 Chromosome translocations of FoxP1 have been detected in the subset of diffused large β-cell lymphomas,12,13 whereas mutations in FoxP3 were associated with the suppression of T-cell immunity that contributed to the growth of ovarian carcinomas.14 GATA3-dependent FoxA1 loss was observed in human breast cancers.15 FoxC2 is overexpressed in invasive breast carcinomas and its increased expression was associated with several adverse prognostic markers, including estrogen receptor negativity and high tumor grade.16 FoxL2 mutation was found in granulosa-cell tumors of the ovary.17 Increased expression of FoxM1 and amplifications of the FoxM1 gene locus were found in various human cancers, including prostate adenocarcinomas, non-small cell lung cancers, glioblastomas, breast adenocarcinomas, and head and neck squamous cell carcinomas.18–22
Based on these studies, Fox proteins provide novel targets for clinical diagnoses and treatment of various human cancers and congenital disorders. In this review, we will focus on FoxM1 transcription factor (previously known as HFH-11B, Trident, Win or MPP2) and summarize recent transgenic and gene knockout studies highlighting the FoxM1 functions in different cell types and tissues in vivo.
FoxM1 is Critical for DNA Replication and Mitosis
FoxM1 was first identified as a proliferation-specific transcription factor, which is expressed in various tumor cell lines and embryonic tissues.23–25 Three isoforms of FoxM1 protein have been described: FoxM1b and FoxM1c function as transcriptional activators, whereas FoxM1a is transcriptionally inactive.23 Multiple in vitro approaches have been used to examine the role of FoxM1 in cellular proliferation. These included FoxM1 mRNA targeting by siRNA, inhibition of FoxM1 by peptides or chemical inhibitors, analysis of a cell line with Doxycycline (Dox)-inducible overexpression of Fox M1b, and analysis of mouse embryonic fibroblasts (MEFs) from Foxm1-/- embryos that lacked all FoxM1 isoforms.26–36 These published studies convincingly demonstrated that FoxM1b and FoxM1c are important regulators of cellular proliferation. FoxM1b induces expression of cyclin A2, JNK1, ATF2 and Cdc25A phosphatase, all of which are critical for G1/S transition and DNA replication.18,30,37 In addition, FoxM1b induces transcription of Skp2 and Cks1 which encode subunits of the Skp/Cullin-1/F-box (SCF) ubiquitin ligase complex, which is required for degradation of Cdk2 inhibitors p21Cip1 and p27Kip1 during G1 phase of the cell cycle.30 Thus, FoxM1b negatively regulates the stability of p21Cip1 and p27Kip1 proteins, leading to increased Cdk2 activity and promoting G1/S transition.
Progression into mitosis requires activation of Cdk1 through the removal of inhibitory phosphates at Thr 14 and Tyr 15 by the Cdc25B and Cdc25C phosphatases.38–40 FoxM1b inactivation resulted in reduced expression of Cdc25B and delayed accumulation of cyclin B1, inhibiting cyclin B-cdk1 kinase activation and entry into mitosis.18 Both Cdc25B and cyclin B1 are direct transcriptional targets of FoxM1b.18 In addition, FoxM1b is a transcriptional activator of various genes critical for chromosome segregation and cytokinesis, such as Aurora B kinase and polo-like kinase 1 (Plk-1), Survivin, Cenp A, B and F isoforms.29,30 Depletion of FoxM1b caused chromosome instability and frequent failure of cytokinesis.29 Altogether, these published studies demonstrated that FoxM1b regulates expression of proteins that are required for DNA replication and mitosis.
FoxM1 Expression during Embryogenesis and Organ Injury
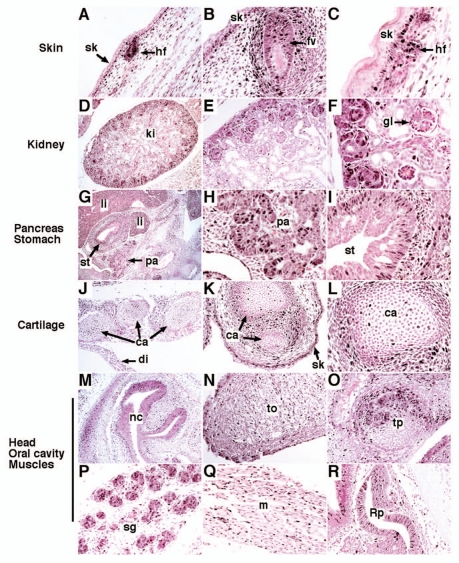
Previous studies used in situ hybridization and immunohistochemistry to demonstrate that FoxM1 is expressed in various cell types during embryogenesis.23,41,42 These include hepatoblasts, cardiomyocytes, smooth muscle and endothelial cells, pancreatic endocrine cells, thymocytes, precursors of granule neurons, as well as epithelium and mesenchyme of the embryonic lung, intestine and liver. While recent studies focused on the role of FoxM1 in these cell types, FoxM1 expression pattern in mouse embryos appears to be much broader. We used immunostaining of embryonic paraffin sections with antibodies against the N-terminal region of the mouse FoxM1 protein43 to identify additional sites of FoxM1 expression in mouse embryos. In developing skin, FoxM1 protein was observed in hair primodiums and keratinocytes of basal layers at embryonic day (E) 15.5 and E18.5 (Fig. 1A–C). FoxM1 was also observed in developing kidney (Fig. 1D–F) and cartilage (Fig. 1J–L), as well as in epithelial and mesenchymal cells of pancreas and stomach (Fig. 1G–I). Several additional FoxM1 expressing regions were found in nasal and oral cavities, tongue, tooth primordiums, salivary glands, muscles, as well as in the Rathke's pouch (Fig. 1M–R), the latter of which gives a rise to pituitary gland. FoxM1 functions in these tissues remain uncharacterized.
Figure 1.
FoxM1 expression in the mouse embryo. Paraffin sections were prepared from mouse wild type embryos harvested at E13.5 (G–I and R), E15.5 (A, B, D and J–P) or E18.5 (C, E, F and Q). Sections were used for immunohistochemistry with FoxM1 antibodies (brown) and counterstained with nuclear fast red (red nuclei). Nuclear FoxM1 staining was detected in hair follicles (A and C), follicles of vibrissae (B) and keratinocytes of basal layers (A–C). FoxM1 was also observed in developing kidney (D–F), cartilage (J–L), epithelial and mesenchymal cells of pancreas and stomach (G and I). FoxM1 expressing cells were detected in nasal cavity (M), tongue (N), tooth primordium (O), salivary glands (P), muscles (Q) and the Rathke's pouch (R). sk, skin; hf, hair follicle; fv, follicle of vibrissae; ki, kidney; gl, glomeruli; li, liver; pa, pancreas; st, stomach; ca, cartilage; di, diaphragm; nc, nasal cavity; to, tongue; tp, tooth primordium; sg, salivary gland; m, muscle; Rp, Rathke's pouch. Magnification: (D, G, J and M) x50; (C, F and L) x400; (A, E and N–P) x100; remaining parts, x200.
While FoxM1 expression in adult mice is restricted to intestinal crypts, thymus and testes,23,24,41 FoxM1 expression is re-activated after organ injury and cancer formation. After lung injury with either butylated hydroxytoluene (BHT) or inflammatory mediator lipopolysaccharide (LPS), FoxM1 protein was found in pulmonary epithelial, endothelial and smooth muscle cells.44,45 FoxM1 expression was induced in hepatocytes and pancreatic endocrine cells following partial hepatectomy or pancreatectomy, respectively.46,47 Following liver injury with carbon tetrachloride (CCl4), FoxM1 expression was induced in hepatocytes, bile duct epithelial cells, and inflammatory cells infiltrating periportal liver regions.43,48 Consistent with the important role of FoxM1 in cell cycle progression, increased levels of FoxM1 mRNA and protein were found in various mouse and human tumors.10,18,19,49 Although FoxM1 expression patterns appear to be consistent with cellular proliferation, subsequent studies showed that FoxM1 functions are diverse and tissue-specific.
Foxm1-/- Mice is Embryonic Lethal
Mice homozygous for Foxm1 null mutation have been previously generated and characterized by two separate labs26,50 (Table 1). Mice with targeting insertion of PGK-neomycin cassette into the third exon of FoxM1 gene (Foxm1-/-neo mice) died immediately after birth due to structural abnormalities in the heart and liver.50 More severe phenotype was observed in a distinct Foxm1-/- mouse line, which contained a complete deletion of exons 4 to 7 that encode the FoxM1 Winged Helix DNA binding and the C-terminal transcriptional activation domains.26 The Foxm1-/- embryos died in utero between E13.5 and E16.5 due to multiple abnormalities in various organ systems, including liver, lungs, blood vessels and heart.26,51,52 Abnormal accumulation of polyploid cardiomyocytes and hepatoblasts, resulting from a failure to complete mitosis, was found in both Foxm1-/-neo and Foxm1-/- mouse lines.26,50–52 Surprisingly, the polyploid phenotype was restricted to the developing heart, liver and muscle layers of blood vessels, whereas other cell types in Foxm1-/- embryos shown no visible changes in either cell sizes or proliferation rates, raising a possibility that FoxM1 functions depend on cellular specificity. These studies demonstrated that FoxM1 is required for proliferation of cardiomyocytes, hepatoblasts and smooth muscle cells during embryogenesis, whereas FoxM1 appears to be dispensable for cellular proliferation in other embryonic cell lineages.
Table 1.
Transgenic mouse models showing the role of FoxM1 in embryonic development
| Mouse model | Affected cells | Phenotypes and references |
| Foxm1-/-neo | Gene disruption in all cell types | Postnatal lethality, accumulation of polyploid cardiomyocytes and hepatoblasts.50 |
| Foxm1-/- | Deletion in all cell types | Embryonic lethality between E13.6 and E17.5, diminished numbers of hepatocytes, absence of intrahepatic bile ducts.26 Diminished proliferation of cardiomyocytes and ventricular hypoplasia in the heart.52 Defects in differentiation of pulmonary mesenchyme into mature capillary endothelial cells, structural defects in blood vessels.51 |
| smMHC-Cre Foxm1fl/fl | Deletion in smooth muscle cells | Postnatal lethality due to severe pulmonary hemorrhage and structural defects in arterial walls and esophagus, diminished proliferation of smooth muscle cells.42 |
| AFPp-Cre Foxm1fl/fl | Deletion in hepatoblasts | Embryonic lethality by E18.5, absence of intrahepatic bile ducts, diminished numbers of hepatoblasts resulting from diminished DNA replication and a failure to enter mitosis causing a polyploid phenotype.26 |
| Math1-Cre Foxm1fl/fl and Nestin-Cre Foxm1fl/fl | Deletion in neural precursor cells | Mice are viable. Entry into mitosis was delayed. Cerebellar granule neuron precursors displayed defects in spindle assembly and centrosome amplification. Differentiation of neuron precursors was normal54 |
| SPC-rtTA/TetO-Cre Foxm1fl/fl | Deletion in respiratory epithelial cells | Postnatal lethality due to respiratory failure after birth, delayed lung maturation, decreased surfactant production. Proliferation of non-mature respiratory epithelial cells was normal.41 |
| Pdx1-Cre Foxm1fl/fl | Deletion in pancreatic epithelial cells | Mice are viable, delayed postnatal β-cell mass expansion, impaired islet function and diabetes in male mice by 9 week of age.56 Gestational diabetes was observed in female mice during pregnancy57 |
| lck-Cre Foxm1fl/fl and CD4-Cre Foxm1fl/fl | Deletion in T lymphocytes | Mice are viable, decreased proliferation of early thymocytes and activated mature T cells, no effects on apoptosis or T cell differentiation55 |
| SPC-rtTA/TetO-FoxM1 activated mutant | Overexpression in respiratory epithelial cells | Mice are viable, decreased lung sacculation in embryos, increased proliferation of respiratory epithelial cells, focal airway hyperplasia resulted from overgrowth of Clara cells.74 |
Conditional Inactivation of FoxM1 during Embryogenesis
Given the broad expression pattern of FoxM1 during embryogenesis,23,41,42 several recent studies used conditional knockout mouse models to address cell-specific roles of FoxM1 in different cell types (Table 1). FoxM1-floxed (fl) mice, containing LoxP sites flanking exons 4 to 7 of the FoxM1 gene, were generated in Robert Costa's laboratory.53 To inactivate the FoxM1 gene in a cell-restricted manner, Foxm1fl/fl mice were bred with various mouse lines containing cell-specific Cre recombinase transgenes. Hepatoblast-specific deletion of FoxM1 in AFPp-Cre Foxm1fl/fl embryos caused embryonic lethality by E18.5 due to disruption of hepatic cords and vasculature, and lack of intrahepatic bile ducts, indicating a cell-autonomous role of FoxM1 in hepatoblasts.26 A conditional deletion of FoxM1 from precursors of cerebellar granule neurons (Math1-Cre and Nestin-Cre transgenes) caused a delay in brain development by interfering with Shh-induced signaling.54 Deletion of FoxM1 from T lymphocyte lineage using either lck-Cre or CD4-Cre transgenes caused a decrease in proliferation of early thymocytes and activated mature T cells without any effects on apoptosis or T-cell differentiation.55 Mice with either endothelial-specific (Tie2-Cre Foxm1fl/fl) or macrophage-specific FoxM1 deletion (mLyz-Cre Foxm1fl/fl) developed normally,43,45 indicating that FoxM1 is not required in these cell lineages during embryonic development. Interestingly, although embryos with pancreatic-specific deletion of FoxM1 (Pdx1-Cre Foxm1fl/fl) displayed normal development of pancreas,56 the male Pdx1-Cre Foxm1fl/fl mice developed severe abnormalities in postnatal β-cell mass expansion, causing impaired islet function and diabetes.56 Female Pdx1-Cre Foxm1fl/fl mice developed gestational diabetes during pregnancy.57 These results are consistent with different requirements for FoxM1 at different stages of pancreatic development.
Recently, we generated transgenic mice in which FoxM1-floxed allele was conditionally deleted using Cre recombinase transgene driven by a smooth muscle myosin heavy chain promoter (smMHC-Cre Foxm1fl/fl mice).42 Although FoxM1 deletion did not influence differentiation of smooth muscle cells, the majority of smMHC-Cre Foxm1fl/fl mice died immediately after birth due to severe pulmonary hemorrhage, structural defects in arterial wall and esophageal abnormalities.42 Reduced levels of cell cycle regulatory genes, such as cyclin B1, Cdc25b, Plk1, JNK1 and cMyc, were found in muscle layers of smMHC-Cre Foxm1fl/fl arteries, as well as in cultured arterial smooth muscle cells after siRNA-mediated FoxM1 depletion.42 Interestingly, decreased myocyte proliferation was specifically found in muscle layers of embryonic blood vessels and esophagus, whereas proliferation rates in either intestinal or peribronchial smooth muscle cells were normal. These results suggest different requirements for FoxM1 in different populations of developing smooth muscle cells. Previously published studies demonstrated that vascular and intestinal smooth muscle cells use distinct transcription factor complexes to activate smooth muscle-specific gene promoters. For example, the Serum response factor (SRF), GATA-5 and Foxf1 transcription factors displayed fundamental differences in either expression pattern or DNA-binding properties between different smooth muscle subtypes.58–61 Although, molecular mechanisms underlying the FoxM1 selectivity in smooth muscle cells remain unknown, it is possible that FoxM1 cooperates with other transcriptional co-activators to regulate different set of genes in subpopulations of smooth muscle cells.
In our recent studies,41 FoxM1 was deleted conditionally in developing respiratory epithelium using the surfactant-associated protein C (SPC) promoter (SPC-rtTA/TetO-Cre Foxm1fl/fl mice). Deletion of FoxM1 did not alter proliferation rates in respiratory epithelial cells, indicating that FoxM1 is not required for epithelial proliferation during lung development. However, the FoxM1 deletion impaired lung maturation and caused respiratory failure after birth.41 Maturation defects in SPC-rtTA/TetO-Cre Foxm1fl/fl lungs were associated with decreased expression of surfactant-associated proteins SPC, SPB and SPA, and a delay in differentiation of type I cells from their epithelial precursors.41 Thus, FoxM1 is dispensable for epithelial proliferation but is critical for surfactant homeostasis and lung maturation during lung development. Altogether, various conditional knockout models demonstrated that FoxM1 plays unique roles in different tissues during embryonic development.
FoxM1 in Organ Injury and Regeneration
Since FoxM1 expression is re-activated in various organ injury models, several recent studies addressed the role of FoxM1 in organ injury and repair (Table 2). Following partial hepatectomy, mice with hepatocyte-specific FoxM1 deletion (Albumin-Cre Foxm1fl/fl) showed reduced DNA replication, which was due to reduced expression of Cdc25A phosphatase and increased nuclear accumulation of p21Cip1 protein in regenerating hepatocytes, causing decreases in Cdk2 activation and hepatocyte progression into S phase.53 Likewise, a significant reduction in numbers of hepatocytes undergoing mitosis was found in regenerating Albumin-Cre Foxm1fl/fl livers. Mitotic abnormalities in Albumin-Cre Foxm1fl/fl livers were due to diminished mRNA levels and nuclear expression of Cdc25B phosphatase and delayed accumulation of cyclin B1, causing a decrease in Cdk1 activation.53 Consistent with the important role of FoxM1 in hepatocyte proliferation, overexpression of FoxM1 in hepatocytes of TTR-FoxM1b transgenic mice reduced p21Cip1 expression and increased proliferation of hepatocytes after partial hepatectomy46 or CCl4-mediated liver injury.48 These results convincingly showed that FoxM1 plays a key role in proliferation of hepatocytes during liver regeneration. Interestingly, hepatocytes of old TTR-FoxM1b mice were protected against age-associated decline in liver regeneration, which normally occurred in regenerating livers of old wild-type mice.62 Increased regenerating potential in old TTR-FoxM1b livers suggests that FoxM1 can be used as an attractive therapeutic target to restore age-associated decline in liver regeneration by inducing hepatocyte proliferation.
Table 2.
Transgenic mouse models showing the role of FoxM1 in organ injury
| Mouse model | Affected cells | Injury/organ | Phenotype |
| Albumin-Cre Foxm1fl/fl | Deletion in hepatocytes | Partial hepatectomy/Livers | Mice showed diminished proliferation of hepatocytes after partial hepatectomy53 |
| Tie2-Cre Foxm1fl/fl | Deletion in endothelial cells | LPS treatment/Lungs | Reduced vascular repair, decreased proliferation of endothelial cells and impaired reannealing of endothelial adherens junctions were found.45,78 |
| Pdx1-Cre Foxm1fl/fl | Deletion in pancreatic epithelial cells | Partial pancreatectomy/Pancreas | Mice showed decreased proliferation of alpha- and beta-cells following partial pancreatectomy. No differences in proliferation of acinal or ductal cells were found.47 |
| Pregnancy/Pancreas | Gestational diabetes was observed in female mice during pregnancy57 | ||
| mLyz-Cre Foxm1fl/fl | Deletion in macrophages and granulocytes | Carbon tetrachloride/Liver | No defects were found in macrophages and granulocytes of bone marrow and peripheral blood. Following acute liver injury by carbon tetrachloride, a delay in liver repair was found. Mice showed decreased recruitment of monocytes into the injured liver, as well as impaired differentiation of monocytes toward mature macrophage lineage.43 |
| Rosa26-FoxM1b | Overexpression in all cell types | BHT treatment/Lungs | Increased proliferation of respiratory epithelial, endothelial and smooth muscle cells was found after BHT-mediated lung injury.44 |
| TTR-FoxM1b | Overexpression in hepatocytes | Partial hepatectomy or carbon tetrachloride/Liver | Increased proliferation of hepatocytes was found after partial hepatectomy46 or carbon tetrachloride-mediated liver injury.48 |
| Partial hepatectomy and aging/Liver | Expression of FoxM1b resulted in protection of old mice against age-associated decline in liver regeneration.62 |
The importance of FoxM1 in pancreatic regeneration was recently found.47 After partial pancreatectomy, Pdx1-Cre Foxm1fl/fl mice exhibited specific impairments in β-cell mass regeneration and islet growth. Although cellular proliferation was reduced in pancreatic α- and β-endocrine cells, no impairments were found in proliferation of acinar or ductal cells.47 Although this study suggests different requirements for FoxM1 in different populations of regenerating pancreatic cells, molecular mechanisms for this selectivity remain unknown.
Mice with endothelial-specific FoxM1 deletion (Tie2-Cre Foxm1fl/fl) displayed significantly increased lung vascular permeability and increased mortality in response to LPS-mediated injury.45 These complex abnormalities in endothelial repair were due to inability of FoxM1-deficient endothelium to restore endothelial barrier function injured by LPS. FoxM1 directly induced expression of β-catenin protein and was critical for reannealing of endothelial adherens junctions after lung injury. Furthermore, gain-of-function studies with Rosa26-FoxM1b transgenic mice confirmed a critical role of FoxM1 in lung repair. After BHT-mediated lung injury, increased proliferation of alveolar endothelial cells and other respiratory cell types was found in Rosa26-FoxM1b mice.44
Recently, we generated transgenic mice with FoxM1 deletion from myeloid cell lineage (mLyz-Cre Foxm1fl/fl), which includes macrophages, monocytes, neutrophils and their precursors. A significant delay in liver repair was found in mLyz-Cre Foxm1fl/fl mice after CCl4-mediated liver injury.43 Surprisingly, FoxM1-deficiency did not influence proliferation of myeloid cells during liver repair, but dramatically reduced recruitment of macrophages and their precursors, monocytes, into injured livers. Expression of L-selectin and the CCR2 chemokine receptor, both critical for monocyte differentiation and recruitment to injured tissues, was decreased in mLyz-Cre Foxm1fl/fl mice.43 In cotransfection experiments, FoxM1 directly induced transcriptional activity of the mouse CCR2 promoter, indicating that CCR2 is a direct transcriptional target of FoxM1. Furthermore, FoxM1 expression in myeloid cells was critical for liver injury because adoptive transfer of wild type monocytes to injured mLyz-Cre Foxm1fl/fl mice restored liver repair and rescued liver function after CCl4 injury.43 These studies demonstrated that FoxM1 is critical for liver repair and required for recruitment of monocytes to the injured liver.
Role of FoxM1 in Carcinogenesis
Since FoxM1 is re-activated during benign and malignant transformations, this protein was extensively studied in various tumor cell lines as well as in tumor tissues obtained from cancer patients. Microarray analysis of human solid tumors demonstrated that FoxM1 is one of the most common overexpressed genes.63 FoxM1 was overexpressed in human non-small cell lung cancers (NSCLC), head and neck squamous carcinomas, hepatocellular carcinomas (HCC), intrahepatic cholangiocarcinomas, colon carcinomas, basal cell carcinomas, infiltrating ductal breast carcinomas, anaplastic astrocytomas, glioblastomas, pancreatic carcinomas, gastric cancer, acute myeloid leukemia and other human tumors and human neoplastic cell lines.10,22,28,49,64–68 Our previous studies have demonstrated that elevated levels of FoxM1 correlated with high proliferation rates in human prostate adenocarcinomas19 and NSCLC.22 Positive correlation was also found between FoxM1 overexpression and increased angiogenesis in human glioblastomas.69 In breast cancer patient samples, FoxM1 levels strongly associated with expression of estrogen receptor alpha.70 FoxM1 was overexpressed in HCC from patients that responded poorly to treatment.64
To study the role of FoxM1 during carcinogenesis, various genetic mouse models were generated (Table 3). We previously developed a transgenic Rosa26-Foxm1 mouse line in which FoxM1b levels were increased in all cell types.44 Ubiquitous overexpression of FoxM1b elevated proliferation of lung tumor cells and increased the number and size of lung tumors induced by tobacco smoke derived carcinogen 3-methylcholanthrene (MCA) and promoted by butylated hydroxytoluene (BHT).71 Likewise, an increase in the number and size of colorectal tumors was found in Rosa26-Foxm1 mice treated with azoxymethane (AOM) and dextran sodium sulfate (DSS).66 FoxM1b transgene cooperated with SV 40 T Antigen to accelerate initiation and progression of prostate adenocarcinomas in Rosa26-Foxm1/TRAMP and Rosa26-Foxm1/LADY double transgenic mice.19 Deletion of FoxM1 from all cell types in Mx-Cre/Foxm1fl/fl mice decreased urethane-mediated lung tumorigenesis22 and delayed the growth and progression of hepatocellular carcinoma (HCC) induced by DEN/phenobarbital treatment.72 Although these studies suggested a critical role for FoxM1 in lung, prostate, colon and liver tumorigenesis, specific requirements for the FoxM1 in different cell populations were not addressed.
Table 3.
Transgenic mouse models showing the role of FoxM1 in tumorigenesis
| Mouse model | Affected cells | Tumor induction | Type of carcinogenesis | Phenotype |
| Rosa26-FoxM1b | Overexpression in all cell types | MCA/BHT | Lung adenomas | Increased numbers and sizes of lung adenomas and increased proliferation of tumor cells were found. FoxM1b induced chronic lung inflammation with macrophage infiltration and lung remodeling.71 |
| AOM/DSS | Colon adenocarcinomas | Increase in the number and size of colorectal tumors and increased proliferation of tumor cells were found.66 | ||
| Breeding with TRA MP or LADY transgenic mice | Prostate adenocarcinomas | FoxM1b cooperated with SV 40 T Antigen to accelerate development of prostate adenocarcinomas in TRAMP and LADY transgenic mice.19 | ||
| TTR-FoxM1b | Overexpression in hepatocytes | DEN/phenobarbital | Hepatocellular carcinoma (HCC) | Increased proliferation of hepatocytes in preneoplastic regions, but no effects on progression of HCC was found.73 |
| SPC-rtTA/TetO-FoxM1 activated mutant | Overexpression in respiratory epithelial cells | Breeding with K-Ras transgenic mice | Lung adenocarcinomas | Expression of FoxM1 mutant alone was insufficient to induce lung adenocarcinomas. However, the FoxM1 transgene cooperated with activated K-Ras to accelerate lung tumor growth in vivo.74 |
| Mx-Cre Foxm1fl/fl | IFN-inducible deletion in all cell types | Urethane | Lung adenomas | Decreased proliferation of lung tumor cells and reduced numbers and sizes of lung adenomas were found.22 |
| DEN/phenobarbital | HCC | Decreased growth and progression of HCC was found.72 | ||
| Albumin-Cre Foxm1fl/fl | Deletion in hepatocytes | DEN/phenobarbital | HCC | Mice showed diminished proliferation of tumor cells and decreased formation of HCC.28 |
| Villin-Cre Foxm1fl/fl | Deletion in colonic epithelial cells | AOM/DSS | Colon adenocarcinomas | Reduction in the development and growth of colorectal tumors were found.66 |
| SPC-rtTA/TetOCre Foxm1fl/fl | Deletion in respiratory epithelial cells | Urethane or MCA/BHT treatment | Lung adenomas | FoxM1 deletion prior to tumor induction prevented lung tumorigenesis. Deletion of FoxM1 in pre-existed lung tumors delayed tumor growth and decreased tumor cell proliferation.75 |
| Tie2-Cre Foxm1fl/fl | Deletion in endothelial cells | Urethane or MCA/BHT treatment | Lung adenomas | Tumor numbers and sizes were increased. Perivascular infiltration by inflammatory cells was observed. Activity of canonical Wnt signaling was increased in lung tumor cells.79 |
To dissect cell-autonomous roles of FoxM1 in cancer lesions, selective targeting of this transcription factor was performed in vivo. Hepatocyte-specific overexpression of FoxM1b in TTR-FoxM1b transgenic mice was insufficient to induce HCC.73 After DEN/phenobarbital treatment TTR-FoxM1b livers showed increased proliferation of hepatocytes in preneoplastic regions, but no effect on progression of HCC was found.73 In our recent studies, specific expression of activated FoxM1b mutant protein in respiratory epithelial cells (SPC-rtTA/TetO-FoxM1b ΔN mice) caused epithelial hyperplasia but was insufficient to induce lung adenocarcinomas.74 These studies demonstrated that FoxM1b might require a “second hit” to transform differentiated hepatocytes or lung epithelial cells into malignant phenotype. The fact that simultaneous overexpression of FoxM1b-ΔN and activated K-Ras accelerated lung tumor growth in vivo74 provides a direct support for this concept.
FoxM1 was found to be required for growth and expansion of HCC and epithelial tumors. Mice with hepatocyte-specific FoxM1 deletion (Albumin-Cre Foxm1fl/fl) displayed diminished proliferation of tumor cells and decreased formation of HCC after DEN/Phenobarbital induction.28 Likewise, reduced growth of colorectal tumors was found in Villin-Cre Foxm1fl/fl mice treated with AOM/DSS.66 Our recent studies demonstrated that deletion of FoxM1 from lung epithelial cells (SPC-rtTA/tetO-Cre/Foxm1fl/fl transgenic mice) prior to the tumor initiation with urethane or MCA/BHT caused a striking reduction in the number and size of lung adenomas.75 Decreased lung tumorigenesis in SPC-rtTA/tetO-Cre/Foxm1fl/fl mice was associated with diminished proliferation of tumor cells and reduced expression of Topoisomerase-2α (TOPO-2α), a critical regulator of tumor cell proliferation. We also demonstrated that FoxM1 directly bound to and induced transcription of the mouse TOPO-2α promoter region, indicating that TOPO-2α is a direct target of FoxM1 in lung tumor cells.75 Interestingly, a deletion of FoxM1 in preexisting lung tumors dramatically reduced tumor growth in the lung,75 indicating that FoxM1 is a promising target for antitumor therapy in cancer patients.
Lung cancer lesions contain not only tumor cells, but also diverse stromal and inflammatory cells, that create a tumor promoting microenvironment. Vascular endothelial cells provide essential support to the tumor microenvironment, directly contributing to proliferation and progression of tumor cells. Recently, we used mice with endothelial cell-specific FoxM1 deletion (Tie2-Cre/Foxm1fl/fl) to study the role of FoxM1 in tumor-associated blood vessels. Surprisingly, numbers and sizes of lung tumors were increased in Tie2-Cre/Foxm1fl/fl mice treated with urethane or MCA/BHT.79 Thus, FoxM1 may function as a tumor suppressor in tumor-associated blood vessels. In addition, perivascular infiltration by inflammatory cells was elevated and numbers of inflammatory cells in BAL fluid were increased in Tie2-Cre/Foxm1fl/fl lungs. FoxM1 was shown to be a direct transcriptional activator of endothelial-specific genes, such as VEGF receptor-2 (Flk-1) and FoxF1 transcription factor, both of which are critical for lung inflammation.79 Moreover, FoxM1 was directly bound to the promoter region of Sfrp1 gene, a known inhibitor of canonical Wnt/β-catenin signaling. As a consequence of decreased Sfrp1 expression in FoxM1-deficient endothelium, Tie2-Cre/Foxm1fl/fl tumors displayed increased canonical Wnt signaling as was demonstrated by activation of TOPGAL transgene,79 a known reporter for Wnt/β-catenin signaling. Taken together, these in vivo studies suggest that endothelial-specific expression of FoxM1 limits lung inflammation and canonical Wnt signaling in lung epithelial cells, thereby restricting lung tumorigenesis.
FoxM1 Functions are not Limited to Regulation of the Cell Cycle
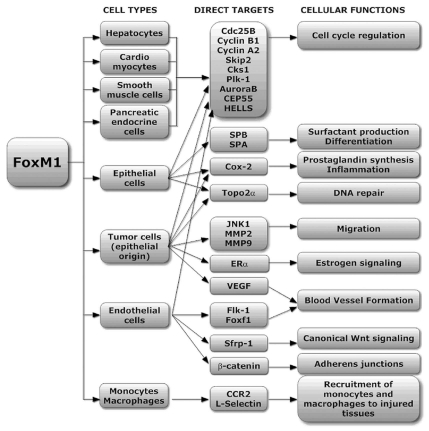
Requirements for FoxM1 in proliferation of tumor cell lines and MEFs in vitro are well described and accepted (reviewed in ref. 18 and 76). However, recent studies demonstrated that FoxM1 deletion in vivo did not always inhibit cellular proliferation. For example, Foxm1-/- embryos survived until late gestation and exhibited several well-developed embryonic tissues without proliferation defects,26 suggesting the role of FoxM1 is cell type specific. Detailed analysis of Foxm1-/- phenotypes led to discovery of additional FoxM1 functions (Fig. 2). FoxM1 was found to be critical for cellular differentiation. For example, livers of Foxm1-/- embryos failed to form intrahepatic bile ducts,26 implicating FoxM1 in differentiation of hepatoblast precursor cells toward the biliary epithelial cell lineage during liver development. Likewise, Foxm1-/- lungs exhibited defects in differentiation of pulmonary mesenchyme into mature capillary endothelial cells during the canalicular stage of lung development.51 Although differentiation of either cardiomyocytes or smooth muscle cells was not altered in FoxM1-deficient embryos,42,52 FoxM1 was required for differentiation of non-mature respiratory epithelial cells toward alveolar type II and type I lineages.41 These results are consistent with different requirements for FoxM1 in various embryonic cell types (Fig. 2).
Figure 2.
Diagram showing direct FoxM1 target genes in different cell lineages.
Lung epithelial-specific deletion of FoxM1 during embryogenesis did not influence epithelial proliferation.41 However, FoxM1 deletion from respiratory epithelium of adult mice inhibited proliferation of lung tumor cells and dramatically decreased lung tumorigenesis induced by either urethane or MCA/BHT.75 Furthermore, FoxM1 was not required for proliferation of pancreatic endocrine cells in utero, but it was found to be critical for proliferation of β-cells after birth.77 These published studies support the hypothesis that the role of FoxM1 may change under various biological conditions.
Additional FoxM1 functions were reported in recent studies. FoxM1 induces cell migration and invasion of cultured osteosarcoma U2OS cells and MEFs through transcriptional activation of the MAPK kinase JNK1 and metalloproteinases MMP-2 and MMP-9.37 Increased angiogenesis in FoxM1-expressing pancreatic adenocarcinoma cell lines was directly linked to the activation of VEGF gene by FoxM1.33 FoxM1 directly binds to and transcriptionally activates the promoter region of Cyclooxygenase 2 (Cox2) and TOPO-2α genes, implicating FoxM1 in prostaglandin synthesis and DNA repair.71,75 In addition, FoxM1 was found to be critical in reannealing of endothelial adherens junctions by directly inducing the β-catenin protein.78 Finally, FoxM1 influenced surfactant homeostasis by activating expression of surfactant-associated proteins SPC, SPB and SPA in respiratory epithelial cells.41 Thus, FoxM1 is important for the execution of distinct cellular functions in various differentiated cells (Fig. 2).
Summary
(1) Although FoxM1 positively regulates cellular proliferation in cultured cell lines in vitro, FoxM1 functions in vivo are diverse and dependent on cellular specificity; (2) FoxM1 is critical for various cellular functions, such as proliferation, differentiation, migration, DNA repair, surfactant production and formation of cellular junctions; (3) FoxM1 functions change dynamically during embryonic development, carcinogenesis and various biological conditions. Identification of novel FoxM1 functions will enable us to determine whether FoxM1 is a promising therapeutic target for various human diseases and cancers.
Acknowledgements
We thank Dr. Craig Bolte for critically reviewing the manuscript. This work was supported by the Research Grant from American Cancer Society, Ohio Division (T.V.K.), the Concern Foundation Grant 84794 (T.V.K.), a DOD New Investigator Award PC080478 (T.V.K.), NIH grants R01 CA142724 (T.V.K.) and R01 HL84151 (V.V.K.), and Research Scholar Grant RSG-06-187-01 from the American Cancer Society, National office (V.V.K.).
References
- 1.Kaestner KH, Lee KH, Schlondorff J, Hiemisch H, Monaghan AP, Schutz G. Six members of the mouse forkhead gene family are developmentally regulated. Proc Natl Acad Sci USA. 1993;90:7628–7631. doi: 10.1073/pnas.90.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 3.Clevidence DE, Overdier DG, Tao W, Qian X, Pani L, Lai E, et al. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc Natl Acad Sci USA. 1993;90:3948–3952. doi: 10.1073/pnas.90.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 5.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 6.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 7.Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunetti-Pierri N, Paciorkowski AR, Ciccone R, Mina ED, Bonaglia MC, Borgatti R, et al. Duplications of FOXG1 in 14q12 are associated with developmental epilepsy, mental retardation and severe speech impairment. Eur J Hum Genet. 2010 doi: 10.1038/ejhg.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 10.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 11.Dong XY, Chen C, Sun X, Guo P, Vessella RL, Wang RX, et al. FOXO1A is a candidate for the 13q14 tumor suppressor gene inhibiting androgen receptor signaling in prostate cancer. Cancer Res. 2006;66:6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- 12.Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large β-cell lymphoma (DLBCL) patients with poor outcome. Blood. 2004;104:2933–2935. doi: 10.1182/blood-2004-03-1209. [DOI] [PubMed] [Google Scholar]
- 13.Haralambieva E, Adam P, Ventura R, Katzenberger T, Kalla J, Holler S, et al. Genetic rearrangement of FOXP1 is predominantly detected in a subset of diffuse large β-cell lymphomas with extranodal presentation. Leukemia. 2006;20:1300–1303. doi: 10.1038/sj.leu.2404244. [DOI] [PubMed] [Google Scholar]
- 14.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 15.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah SP, Kobel M, Senz J, Morin RD, Clarke BA, Wiegand KC, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009;360:2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 18.Costa RH, Kalinichenko VV, Major ML, Raychaudhuri P. New and unexpected: forkhead meets ARF. Curr Opin Genet Dev. 2005;15:42–48. doi: 10.1016/j.gde.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spirin KS, Simpson JF, Takeuchi S, Kawamata N, Miller CW, Koeffler HP. p27/Kip1 mutation found in breast cancer. Cancer Res. 1996;56:2400–2404. [PubMed] [Google Scholar]
- 21.Singh B, Gogineni SK, Sacks PG, Shaha AR, Shah JP, Stoffel A, et al. Molecular cytogenetic characterization of head and neck squamous cell carcinoma and refinement of 3q amplification. Cancer Res. 2001;61:4506–4513. [PubMed] [Google Scholar]
- 22.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The forkhead box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 23.Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao KM, Sha M, Lu Z, Wong GG. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property and alternative splicing within the DNA binding domain. J Biol Chem. 1997;272:19827–19836. doi: 10.1074/jbc.272.32.19827. [DOI] [PubMed] [Google Scholar]
- 26.Krupczak-Hollis K, Wang X, Kalinichenko VV, Gusarova GA, Wang IC, Dennewitz MB, et al. The Mouse Forkhead Box m1 Transcription Factor is Essential for Hepatoblast Mitosis and Development of Intrahepatic Bile Ducts and Vessels during Liver Morphogenesis. Dev Biol. 2004;276:74–88. doi: 10.1016/j.ydbio.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 27.Major ML, Lepe R, Costa RH. Forkhead Box M1B (FoxM1B) Transcriptional Activity Requires Binding of Cdk/Cyclin Complexes for Phosphorylation-Dependent Recruitment of p300/CBP Co-activators. Mol Cell Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalinichenko VV, Major M, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Forkhead box m1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 30.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radhakrishnan SK, Bhat UG, Hughes DE, Wang IC, Costa RH, Gartel AL. Identification of a chemical inhibitor of the oncogenic transcription factor forkhead box M1. Cancer Res. 2006;66:9731–9735. doi: 10.1158/0008-5472.CAN-06-1576. [DOI] [PubMed] [Google Scholar]
- 32.Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM, et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281:25167–25176. doi: 10.1074/jbc.M603906200. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Downregulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–8300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- 34.Li SK, Smith DK, Leung WY, Cheung AM, Lam EW, Dimri GP, et al. FoxM1c counteracts oxidative stress-induced senescence and stimulates Bmi-1 expression. J Biol Chem. 2008;283:16545–16553. doi: 10.1074/jbc.M709604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–4305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Z, Tan G, Ding M, Dong D, Chen T, Meng X, et al. Foxm1 transcription factor is required for maintenance of pluripotency of P19 embryonal carcinoma cells. Nucleic Acids Res. 2010;38:8027–8038. doi: 10.1093/nar/gkq715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang IC, Chen YJ, Hughes DE, Ackerson T, Major ML, Kalinichenko VV, et al. FOXM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J Biol Chem. 2008;283:20770–20778. doi: 10.1074/jbc.M709892200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borgne A, Meijer L. Sequential dephosphorylation of p34(cdc2) on Thr-14 and Tyr-15 at the prophase/metaphase transition. J Biol Chem. 1996;271:27847–27854. doi: 10.1074/jbc.271.44.27847. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson I, Hoffmann I. Cell cycle regulation by the Cdc25 phosphatase family. Prog Cell Cycle Res. 2000;4:107–114. doi: 10.1007/978-1-4615-4253-7_10. [DOI] [PubMed] [Google Scholar]
- 40.Wells NJ, Watanabe N, Tokusumi T, Jiang W, Verdecia MA, Hunter T. The C-terminal domain of the Cdc2 inhibitory kinase Myt1 interacts with Cdc2 complexes and is required for inhibition of G(2)/M progression. J Cell Sci. 1999;112:3361–3371. doi: 10.1242/jcs.112.19.3361. [DOI] [PubMed] [Google Scholar]
- 41.Kalin TV, Wang IC, Meliton L, Zhang Y, Wert SE, Ren X, et al. Forkhead Box m1 transcription factor is required for perinatal lung function. Proc Natl Acad Sci USA. 2008;105:19330–19335. doi: 10.1073/pnas.0806748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ustiyan V, Wang IC, Ren X, Zhang Y, Snyder J, Xu Y, et al. Forkhead box M1 transcriptional factor is required for smooth muscle cells during embryonic development of blood vessels and esophagus. Dev Biol. 2009;336:266–279. doi: 10.1016/j.ydbio.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren X, Zhang Y, Snyder J, Cross ER, Shah TA, Kalin TV, et al. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol. 2010;30:5381–5393. doi: 10.1128/MCB.00876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalinichenko VV, Gusarova GA, Tan Y, Wang IC, Major ML, Wang X, et al. Ubiquitous expression of the forkhead box M1B transgene accelerates proliferation of distinct pulmonary cell-types following lung injury. J Biol Chem. 2003;278:37888–37894. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- 45.Zhao YY, Gao XP, Zhao YD, Mirza MK, Frey RS, Kalinichenko VV, et al. Endothelial cell-restricted disruption of FoxM1 impairs endothelial repair following LPS-induced vascular injury. J Clin Invest. 2006;116:2333–2343. doi: 10.1172/JCI27154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye H, Holterman A, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S-phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ackermann Misfeldt A, Costa RH, Gannon M. Beta-cell proliferation, but not neogenesis, following 60% partial pancreatectomy is impaired in the absence of FoxM1. Diabetes. 2008;57:3069–3077. doi: 10.2337/db08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Hung NJ, Costa RH. Earlier expression of the transcription factor HFH 11B (FOXM1B) Diminishes Induction of p21CIP1/WAF1 levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology. 2001;33:1404–1414. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- 49.Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M, et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS One. 2009;4:4849. doi: 10.1371/journal.pone.0004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korver W, Schilham MW, Moerer P, van den Hoff MJ, Dam K, Lamers WH, et al. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor trident. Curr Biol. 1998;8:1327–1330. doi: 10.1016/s0960-9822(07)00563-5. [DOI] [PubMed] [Google Scholar]
- 51.Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box M1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280:22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- 52.Ramakrishna S, Kim IM, Petrovic V, Malin D, Wang IC, Kalin TV, et al. Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev Dyn. 2007;236:1000–1013. doi: 10.1002/dvdy.21113. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b Transcription Factor is Essential for Hepatocyte DNA Replication and Mitosis during Mouse Liver Regeneration. Proc Natl Acad Sci USA. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, et al. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol. 2007;27:8259–8270. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue L, Chiang L, He B, Zhao YY, Winoto A. FoxM1, a forkhead transcription factor is a master cell cycle regulator for mouse mature T cells but not double positive thymocytes. PloS One. 2010;5:9229. doi: 10.1371/journal.pone.0009229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, et al. The FoxM1 transcription factor is required to maintain pancreatic betacell mass. Mol Endocrinol. 2006;20:1853–1866. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H, Zhang J, Pope CF, Crawford LA, Vasavada RC, Jagasia SM, et al. Gestational diabetes mellitus resulting from impaired beta-cell compensation in the absence of FoxM1, a novel downstream effector of placental lactogen. Diabetes. 2010;59:143–152. doi: 10.2337/db09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manabe I, Owens GK. CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J Clin Invest. 2001;107:823–834. doi: 10.1172/JCI11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lilly B, Olson EN, Beckerle MC. Identification of a CArG box-dependent enhancer within the cysteine-rich protein 1 gene that directs expression in arterial but not venous or visceral smooth muscle cells. Dev Biol. 2001;240:531–547. doi: 10.1006/dbio.2001.0507. [DOI] [PubMed] [Google Scholar]
- 60.Morrisey EE, Ip HS, Tang Z, Lu MM, Parmacek MS. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 61.Kalinichenko VV, Lim L, Beer-Stoltz D, Shin B, Rausa FM, Clark J, et al. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the forkhead box f1 transcription factor. Dev Biol. 2001;235:489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- 62.Wang X, Quail E, Hung NJ, Tan Y, Ye H, Costa RH. Increased Levels of Forkhead Box M1B Transcription Factor in Transgenic Mouse Hepatocytes Prevents Age-Related Proliferation Defects in Regenerating Liver. Proc Natl Acad Sci USA. 2001;98:11468–11473. doi: 10.1073/pnas.201360898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 65.Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 Is a Downstream Target of Gli1 in Basal Cell Carcinomas. Cancer Res. 2002;62:4773–4780. [PubMed] [Google Scholar]
- 66.Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–1431. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 67.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakamura S, Hirano I, Okinaka K, Takemura T, Yokota D, Ono T, et al. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis. 2010;31:2012–2021. doi: 10.1093/carcin/bgq185. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68:8733–8742. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM, et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010;29:2983–2995. doi: 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang IC, Meliton L, Tretiakova M, Costa RH, Kalinichenko VV, Kalin TV. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene. 2008;27:4137–4149. doi: 10.1038/onc.2008.60. [DOI] [PubMed] [Google Scholar]
- 72.Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, et al. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalinina OA, Kalinin SA, Polack EW, Mikaelian I, Panda S, Costa RH, et al. Sustained hepatic expression of FoxM1B in transgenic mice has minimal effects on hepatocellular carcinoma development but increases cell proliferation rates in preneoplastic and early neoplastic lesions. Oncogene. 2003;22:6266–6276. doi: 10.1038/sj.onc.1206640. [DOI] [PubMed] [Google Scholar]
- 74.Wang IC, Zhang Y, Snyder J, Sutherland MJ, Burhans MS, Shannon JM, et al. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Dev Biol. 2010;347:301–314. doi: 10.1016/j.ydbio.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang IC, Meliton L, Ren X, Zhang Y, Balli D, Snyder J, et al. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PloS One. 2009;4:6609. doi: 10.1371/journal.pone.0006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Costa RH, Kalinichenko VV, Holterman AX, Wang X. Transcription factors in liver development, differentiation and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 77.Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, et al. The FoxM1 transcription factor is required to maintain pancreatic betacell mass. Mol Endocrinol. 2006;20:1853–1866. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- 78.Mirza MK, Sun Y, Zhao YD, Potula HH, Frey RS, Vogel SM, et al. FoxM1 regulates re-annealing of endothelial adherens junctions through transcriptional control of beta-catenin expression. J Exp Med. 2010;207:1675–1685. doi: 10.1084/jem.20091857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balli D, Zhang Y, Snyder J, Kalinichenko VV, Kalin TV. Endothelial Cell-specific Deletion of Transcription Factor FOXM1 Increases Urethane-induced Lung Carcinogenesis. Cancer Res. 2011;71:40–50. doi: 10.1158/0008-5472.CAN-10-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]