Abstract
MicroRNAs (miRNAs) have emerged as critical regulators of numerous biological processes by modulating gene expression at the post-transcriptional level. It has become increasingly clear that almost all aspects of skeletal muscle development involve regulation by miRNAs. Many of these miRNAs have distinct expression profiles in skeletal muscles, under the regulation by the myogenic program. In the last few years the field has seen a rapid expansion of our knowledge of myogenic miRNAs that target a wide range of muscle genes to coordinately control the myogenic process. In this review we provide an up-to-date list of reported myogenic miRNAs and survey their expression patterns, regulation of biogenesis and gene targets in skeletal muscles. Emerging themes of miRNA regulation in the context of skeletal myogenesis will also be discussed.
Key words: microRNA, skeletal muscle, myogenesis, myoblast differentiation, muscle regeneration, miRNA expression, miRNA targets, feedback
MicroRNAs (miRNAs) are a class of evolutionally conserved non-coding RNAs of ∼22 nucleotides, which regulate gene expression predominantly at the post-transcriptional level.1 Eight years after the initial discovery of lin-4 in C. elegans in 1993,2,3 dozens of miRNAs were reported simultaneously by several groups,4–6 thus beginning the miRNA era. Over the last decade, hundreds of miRNAs were identified or predicted across a wide spectrum of species.7 The prevalence and importance of this small RNA family in modulating gene expression has become increasingly clear. In this review, we will focus on one particular aspect of miRNA function—regulation of skeletal myogenesis. Intensive research activities in this area in recent years have led to the revelation that almost all aspects of skeletal muscle development involve regulation by miRNAs, with these myogenic miRNAs targeting a wide range of muscle genes to coordinately control the myogenic process. Here we will survey the myogenic miRNAs reported so far, and discuss the regulation of their expression in muscles, their gene targets and functions in skeletal myogenesis, and emerging themes of miRNA regulation.
Introduction: miRNA Biogenesis and Targeting
Mammalian miRNAs are encoded in the intergenic or intragenic (both intronic and exonic) regions of the genome, their genes often found clustered.8 MiRNA genes are transcribed as long primary transcripts (pri-miRNAs), mainly by RNA polymerase II,9 although polymerase III-dependent transcription has also been described in reference 10. Pri-miRNAs are processed co-transcriptionally by the nuclear RNase III Drosha/DGCR8 to generate ∼70 bp pre-miRNAs, which are then exported by Exportin-5 to the cytoplasm in a Ran-GTP dependent manner. In the cytoplasm, pre-miRNAs are further processed by the RNase III Dicer to yield ∼22 nt mature miRNAs (reviewed in ref. 9). The miRNA biogenesis paradigm has recently been expanded to include “mirtrons” that bypass Drosha processing,11,12 and miRNAs (or miRNA-like small RNAs) that are generated independently of Dicer.13–16 In addition, recent discoveries of secreted miRNAs in microvehicles communicating between cells further widen the scope of miRNA biogenesis and function.17
Through partial sequence complementarity, miRNAs exert their functions by binding to the 3'UTR of the target mRNAs18 and subsequently directing them for translational inhibition and mRNA decay,19 although translational activation by miRNAs has also been reported in reference 20 and 21. A most recent study has revealed that mammalian miRNAs inhibit gene expression predominantly through decreasing target mRNA levels.22 It has been estimated that over 60% of the human protein-coding genes are under selective pressure to maintain miRNA sites.23 With this enormous target repertoire, it is not surprising that miRNAs have emerged as key regulators for myriads of cellular and developmental processes. The early embryonic lethality of Dicer deficient mice and severe phenotypes from conditional Dicer knockout during different development stages and in various adult tissues,24–27 attest to the pivotal roles of miRNAs.
Regulation of miRNA Expression in Skeletal Muscle
During skeletal muscle development, cells from the somites commit to myogenic lineage and progress along the myogenic pathway by proliferation, terminal differentiation and formation of multinucleated myofibers.28 The activation of muscle specific transcription factors, including MyoD and MEF2 families of proteins, results in reprogramming of gene expression to govern skeletal myogenesis.29,30 Modulation of myogenic gene expression by miRNAs has emerged as a new level of control for myogenesis. Mice with Dicer deleted in muscle die perinatally and display decreased skeletal muscle mass, increased apoptosis of the muscle cells, accompanied by abnormal myofiber morphology.31 It is important to note that Dicer is also responsible for the production of other types of small RNAs, whereas DGCR8, the binding partner of Drosha, is specific to the miRNA biogenesis pathway.9 Of significance, striated muscle-specific Dgcr8 knockout mice display severe dilated cardiomyopathy and heart failure,32 although characterization of skeletal muscle in those mice has not been reported. Below we survey the expression of miRNAs with reported myogenic functions in skeletal muscle (Table 1).
Table 1.
Expression and targets of miRNAs known to function in skeletal myogenesis
| microRNA | Tissue distribution | Expression upon myoblast differentiation | Targets in skeletal muscles |
| miR-1a | muscle-specific | Increase | HDAC4,40 Cx43,79 Pax7,47 c-Met,80 G6PD84 |
| miR-24 | ubiquitous | Increase | unknown |
| miR-26a | ubiquitous | Increase | Ezh238 |
| miR-27b | ubiquitous | Increase | Pax359 |
| miR-29b/c | ubiquitous | Increase | YY1,54 COL1A1, ELN84,94 |
| miR-125b | ubiquitous | Decrease | IGF-II66 |
| miR-133 | muscle-specific | Increase | SRF,40 nPTB,87 UCP288 |
| miR-181 | ubiquitous | Increase | Hox-A1158 |
| miR-206a | skeletal muscle-specific | Increase | DNA polα,78 Fstl1,35 Utrn,35 Cx43,79 TIMP3,86 Pax7,47,52,84 c-Met,80,81 HDAC482 |
| miR-208b/499 | muscle-specific | Increase | Sox6,49 Purβ,49 Sp3,49 HP-1β49 |
| miR-214 | ubiquitous | Increase | Ezh2,53 N-Ras101 |
| miR-221/222 | ubiquitous | Decrease | p2761 |
| miR-322/424 | ubiquitous | Increase | Cdc25A60 |
| miR-486 | muscle-enriched | Increase | FoxO1,51 PTEN,51 Pax752 |
| miR-503 | ubiquitous | Increase | Cdc25A60 |
miR-1 and miR-206 presumably share the same targets. Here we list the targets separately based on published experimental evidence.
MiR-1/miR-133/miR-206.
The best-studied myogenic miRNAs are the miR-1/miR-206 and miR-133a/miR-133b families, consisting of four mature miRNAs expressed from three chromosomal loci as bicistronic transcripts.33 These miRNAs are specifically expressed in cardiac and skeletal muscles under the control of the myogenic transcription factors SRF, MyoD and MEF2,34–37 and they regulate the fundamental processes of skeletal myogenesis including myoblast/satellite cell proliferation and differentiation (reviewed in ref. 33). Among many miRNAs profiled, miR-1, miR-133 and miR-206 levels are most dramatically increased during myoblast differentiation.38–40
Recently, we have reported that miR-1 expression, under the control of its upstream and intragenic enhancers,34,37 is regulated by mammalian target of rapamycin (mTOR) signaling.39 This regulation is mediated by mTOR control of MyoD stability.39 MyoD also regulates the expression of miR-133,37 and miR-206,35 and, as a key myogenic transcription factor, it is placed directly upstream of several other candidate myogenic miRNAs (discussed later). Interestingly, the results of our miRNA profiling suggest that the drastic increase of miR-133 and miR-206 levels during myoblast differentiation, like that of miR-1, is completely inhibited by the mTOR-specific inhibitor rapamycin.39 In addition, several other miRNAs are induced during myogenic differentiation, albeit to lesser degrees, in a rapamycin-sensitive manner.39 Hence, it is an intriguing possibility that the mTOR-MyoD axis may regulate a cohort of miRNAs in the coordinate regulation of skeletal myogenesis.
Adding to the complexity of regulation, miR-1, miR-133 and miR-206 have also been suggested to be controlled at the level of pri-miRNA processing. The RNA binding protein KSRP41 is a component of both Drosha and Dicer complexes and it promotes the maturation of a subset of miRNA precursors through binding to their terminal loops.42 MiR-1, miR-133 and miR-206 appear to belong to this subgroup of miRNAs. Future studies to examine the commonality of this regulatory mechanism in myogenic miRNAs should be illuminating.
The expression of these three well-studied myogenic miRNAs has also been examined in vivo. MiR-1,39,43,44 miR-133,43 and miR-206,43–46 have all been reported to be upregulated during muscle regeneration in animals after injury. Interestingly, a transient drop in these miRNA levels has also been observed immediately after muscle injury.43,44,46,47 Whereas this decrease may be a consequence of muscle damage and induction of fibrosis independent of muscle regeneration,44 a recent report correlated this early phase of miR-1 and miR-206 suppression with their anti-proliferation effect on satellite cells.47 miR-1 and miR-133a levels were also found decreased in skeletal muscles seven days after functional overload, accompanied by an increase in the corresponding pri-miRNAs,48 implicating regulation at a miRNA maturation step. As the authors speculated, potential targets of these miRNAs may be involved in muscle growth regulation.48 Alternatively, it is reasonable to suggest, based on the recent report,47 that satellite cell activation and proliferation in response to the overload stimulus may require suppression of miR-1.
MiR-208/miR-499.
Also displaying a muscle-restricted expression pattern is a family of intronic miRNAs named “MyomiRs,”49 consisting of miR-208a, miR-208b and miR-499. Whereas miR-208a is restricted to cardiac muscle, miR-208b and miR-499 are expressed in both cardiac and skeletal muscles. These miRNAs are encoded by introns of their host myosin genes, α-MHC, β-MHC and Myh7b, respectively, regulating myofiber type specification.49 Interestingly, miR-499 was found to be the most dramatically decreased miRNA in a hind limb suspension-induced atrophy model, suggesting its potential importance in muscle maintenance.50
MiR-486.
Another muscle-enriched miRNA is miR-486. Encoded in the gene Ankrin-1, the expression of miR-486 is activated by myocardin-related transcription factor-A (MRTF-A), SRF and MyoD.51 Most recently, miR-486 is reported to be highly induced during myoblast differentiation, and to exert a myogenic function.52
Ubiquitously expressed miRNAs with myogenic functions.
In addition to the muscle-restricted and muscle-enriched miRNAs described above, some ubiquitously expressed miRNAs also have myogenic functions, and their expression levels typically change during myoblast differentiation. MiR-214 expression is de-repressed from the polycomb protein Ezh2 and stimulated by MyoD and myogenin during differentiation.53 Similarly, miR-29b/c expression is suppressed in myoblasts by the repressive transcription factor YY1; during myoblast differentiation and muscle regeneration, release from YY1 and stimulation by SRF and MEF2 lead to the upregulation of miR-29 levels.54 MiR-24 transcription is also upregulated during myoblast differentiation, upon de-repression from TGFβ-Smad signaling.55 De-repression from suppressive signals appears to be a common theme within this subset of myogenic miRNAs, the upregulation of which correlates with myogenic differentiation.
Several other miRNAs with myogenic functions are also upregulated during skeletal myogenesis, but the upstream regulators are yet to be identified. MiR-181 is highly expressed in neurons and bone marrow but barely detectable in mature myofibers.56,57 However, miR-181 levels are drastically increased during myoblast differentiation and muscle regeneration.58 Other ubiquitously expressed myogenic miRNAs found to be upregulated during myogenic differentiation include miR-26a,38 miR-27b,59 miR-322/424 and miR-503.60
MiRNAs downregulated during myogenic differentiation.
Compared to myogenic miRNAs that are upregulated, fewer miRNAs with demonstrated myogenic functions are downregulated during myogenic differentiation. MiR-221 and miR-222, which share sequence similarity and are clustered on chromosome X, are found to be highly expressed in quail myoblasts, and downregulated during differentiation.61 A similar expression pattern is found in mouse C2C12 myocytes, although the change is to a lesser degree.61 It has been suggested that the expression of miR-221/222 is under the control of Ras-MAPK signaling,61 consistent with a negative role that this pathway plays in myogenic differentiation.62,63 MiR-222 expression has also been correlated with the presence of infiltrating inflammatory cells in injured muscles,44 and it is highly upregulated in several muscular dystrophies.64 Another miRNA downregulated during myoblast differentiation is miR-125b,65,66 a ubiquitously expressed and brain-enriched miRNA. Most recently we have identified a role for miR-125b as a suppressor of myogenic differentiation and muscle regeneration.66 Expression of miR-125b at the transcriptional level is negatively controlled by mTOR signaling in an mTOR kinase-independent manner.66 True to its emerging status of a master regulator, rapamycin-sensitive mTOR signaling regulates the biogenesis of two miRNAs in opposite directions—enhancing miR-1 levels39 and suppressing miR-125b levels, through mTOR kinase-dependent (our unpublished observations) and -independent66 pathways, respectively. Further delineation of the regulatory mechanism of miR-125b biogenesis will require identification of its cis regulatory elements, which is also the case for many of the other myogenically regulated miRNAs mentioned above.
MiRNA expression during skeletal myogenesis has been profiled extensively by numerous groups. Many miRNAs not mentioned above have been found to be differentially expressed during myoblast differentiation in vitro38–40,47,52,67 and muscle development or disease in vivo.44,47,64,68 Future characterization of those miRNAs will likely lead to the identification of novel myogenic regulators.
Gene Targets and Functions of Myogenic miRNAs
Manipulation of miRNA levels in vitro and in vivo, by genetic and biochemical methods, has become a standard approach to interrogating miRNA functions. Ultimately, revealing the gene targets of a miRNA is key to understanding its function. Identification of biologically important targets remains a major challenge in miRNA studies, as most metazoan miRNAs pair with their targets imperfectly. Pairing of miRNA “seed” region (nucleotide 2–8) to the 3'UTR of the target gene is believed to be one of the most important parameters that determine efficient miRNA targeting,69–71 yet “seedless” targeting has also been well documented.72,73 Although computational prediction has become a powerful tool in facilitating miRNA target identification,18,74,75 the current algorithms remain far from accurate, with high false-positive and false-negative rates.76 Experimental approaches to identifying miRNA targets continue to be developed and refined, but extracting biologically important information from typically very large data sets from such methods can be daunting. The reader is referred to Thomas et al.77 for an excellent review on current approaches to miRNA target identification.
Despite the technical challenges, in recent years numerous biological targets for myogenic miRNAs have been revealed, representing a wide range of genes in the myogenic program, from transcription regulators, signaling molecules, to structural proteins. Here we survey the myogenic miRNAs for which gene targets have been identified or reasonably predicted (Table 1).
MiR-1 and miR-206.
miR-1 and miR-206 differ by only four nucleotides outside the seed region, and they are believed to share gene targets although not all reported targets have been experimentally validated for both miRNAs. The gap junction protein connexin43 (Cx43) is a target of miR-1/miR-206,78,79 the downregulation of which is necessary for myoblast fusion.79 The receptor tyrosine kinase c-Met is also targeted by miR-1/miR-206, identified in a rhabdomyosarcoma cell line; miR-1/miR-206 could function as a potent tumor suppressor in c-Met-overexpressing tumors with aberrant proliferation and cell migration.80,81 Another target of miR-1/miR-206 is histone deacetylase 4 (HDAC4),40,82 a transcriptional repressor of muscle gene expression.83 HDAC4 partly mediates the effects of miR-206 in promoting regeneration of neuromuscular synapses and delaying amyotrophic lateral sclerosis (ALS) in a mouse model.82 Recently, we have found the myogenic fusion-promoting factor follistatin to be regulated by miR-1-controlled HDAC4, and uncovered an mTOR-MyoD-miR-1-HDAC4-follistatin pathway that regulates myocyte fusion in skeletal myogenesis.39 Moreover, miR-1 and miR-206 have recently been reported to target Pax7 and, as a consequence, inhibit satellite cell proliferation and promote myogenic differentiation.47,52,84 Interestingly, miR-1 and miR-206 levels undergo a temporary drop during the early phase of muscle regeneration in animals,44,46,47,66 which correlates well with active satellite cell proliferation at that stage.85
Several more gene targets have been reported for miR-206, although it remains to be determined whether they are also shared by miR-1. Examining the effect of miR-206 overexpression on mRNA expression profiles in C2C12 myoblasts led to the identification of DNA polymerase alpha as a bona fide target of miR-206,78 the downregulation of which may be involved in the suppression of DNA synthesis upon myogenic differentiation. Butyrate-induced transcript 1 (B-ind1), monocyte-to-macrophage differentiation-associated protein (Mmd), and Cx43 were also implicated as miR-206 targets in the same study.78 As discussed earlier, Cx43 has been identified as a target for both miR-1 and miR-206 in another study.79 More targets of miR-206 have been computationally predicted and experimentally confirmed, including follistatin-like 1 (Fstl1),35 utrophin (Utrn),35 and tissue inhibitor of metalloproteinase 3 (TIMP3).86 Glucose-6-phosphate dehydrogenase (G6PD) has been reported to be a miR-1 target; in Duchenne muscular dystrophy the dystrophic phenotype is partially caused by HDAC2 suppression of miR-1 expression and subsequent G6PD over-production.84
MiR-133.
Its genes clustering with those of miR-1 and miR-206, miR-133 targets SRF, thus enhancing myoblast proliferation and inhibiting myogenic differentiation.40 Interestingly, miR-133 has also been shown to directly promote differentiation by targeting the alternative splicing factor neuronal polypyrimidine tract-binding protein (nPTB), the downregulation of which is necessary for the production of muscle-specific transcripts during myogenic differentiation.87 MiR-133 is also reported to target uncoupling protein 2 (UCP2) known to regulate energy expenditure and thermogenesis in various organisms and to negatively impact myoblast differentiation.88
MiR-24.
Found to be suppressed by TGFβ signaling, miR-24 promotes myoblast differentiation.55 The targets of miR-24 in myogenesis have not been reported. Interestingly, in non-myogenic cells miR-24 has been found to target a cohort of cell cycle regulators, including Myc and E2F.72 Hence, it is conceivable that miR-24 may target these same genes in myoblasts and support cell cycle exit necessary for myogenic differentiation.
MiR-26a.
Upregulated during myoblast differentiation, miR-26a has been shown to promote differentiation.38 The histone methyltransferase Ezh2, a polycomb group protein known to negatively regulate skeletal myogenesis,89 is found to be a target of miR-26a.38 Ezh2 binds to chromatin via its association with the transcription factor YY1, and subsequently silences muscle gene expression through its histone methyltransferase activity.89 Hence, miR-26a suppression of Ezh2 may be necessary for myogenic differentiation. Dysregulated expression of miR-26a and Ezh2 has been found in rhabdomyosarcoma, implicating their involvement in the pathogenesis of this malignant tumor.90
MiR-27b.
In a search for miRNAs that target Pax3, miR-27b emerged as a strong candidate predicted by multiple computational algorithms, and a direct targeting was confirmed experimentally.59 Pax3 is required for muscle stem cell maintenance and migration, but its removal is necessary for myogenic differentiation.91,92 MiR-27b promotes entry into the differentiation program both in vitro and in regenerating muscles by downregulating Pax3.59
MiR-29.
MiR-29 targets the polycomb group protein associated transcription factor YY1.54 YY1 recruits PcG proteins to suppress muscle gene expression, and is downregulated by NFκB transcriptionally93 and by miR-29 post-transcriptionally54 to allow myogenic differentiation. In addition, miR-29 targets collagen (COL1A1) and elastin (ELN) in the extracellular matrix in cardiac myocytes,94 and the same targeting has been suggested to mediate the fibrosis phenotype in Duchenne muscular dystrophy due to suppression of miR-29.84
MiR-125b.
As one of the few miRNAs downregulated during myogenic differentiation,65,66 miR-125b is found to negatively regulate myoblast differentiation and muscle regeneration by targeting IGF-II,66 a critical inducer of skeletal myogenesis.95 As discussed earlier, mTOR signaling regulates miR-125b biogenesis in this context.66 We have also reported that the transcription of IGF-II in differentiating myoblasts and regenerating muscles is regulated by mTOR.96,97 Thus, mTOR controls myogenic IGF-II production at both transcriptional and post-transcriptional levels, highlighting the critical role of the mTOR-IGF-II axis in skeletal myogenesis. Other gene targets have been reported for miR-125b in non-myogenic cells, including Lin-28,98 and p53.99 Whether these genes are targeted by miR-125b in muscle and whether miR-125b targets IGF-II in non-muscle cells are yet to be determined.
MiR-181.
The homeobox protein Hox-A11, reported to repress transcription of MyoD,100 is found to be a target for miR-181.58 The transient expression of miR-181 during myoblast differentiation and muscle regeneration, and the lack of its detection in adult muscles, suggest that this microRNA is involved in muscle formation but not maintenance.58
MiR-208b and miR-499.
Produced from introns of two myosin genes—β-MHC and Myh7b, miR-208b and miR-499 share the same seed sequence and presumably the same set of targets. These two miRNAs function redundantly to target the transcriptional repressors of slow myofiber genes, including Sox6, Purβ, Sp3 and HP-1β, thus governing myofiber type switch.49
MiR-214.
Like miR-26a, miR-214 also targets Ezh2,53 a repressor of muscle gene expression. In addition, N-Ras has been found to be a target of miR-214 through transcriptional profiling of miR-214-overexpressing C2C12 myoblasts.101 Since Ras promotes cell cycle entry and inhibits differentiation, miR-214 suppression of N-Ras facilitates cell cycle exit and subsequent myogenic differentiation.101 In zebrafish, miR-214 is involved in muscle fate determination by modulating Hedgehog signaling.102 It will be interesting to see whether this function is conserved in mammals.
MiR-221 and miR-222.
MiR-221 and miR-222 target the cell cycle inhibitor p27Kip in cancer cells.103,104 The same targeting has been implicated in myoblasts, and miR-221/miR-222 inhibits myogenic differentiation possibly through interfering with cell cycle withdrawal involving p27.61 It has been suggested that p27 may also control myoblast differentiation through other cellular targets beyond cell cycle regulation.105
MiR-322/424 and miR-503.
MiR-322/424 (miR-424 is the human ortholog of mouse miR-322) and miR-503 have very similar seed sequences, and their genes are clustered. These two miRNAs are reported to target Cdc25A,60 a phosphatase responsible for removing inhibitory phosphorylation of Cdk2. Upregulation of miR-322/424 and miR-503 in differentiating C2C12 cells leads to cell cycle withdrawal through downregulation of Cdc25A, thus promoting myogenesis.60
MiR-486.
Found to be enriched in cardiac and skeletal muscles, miR-486 was reported to target two negative regulators in the PI3K pathway—PTEN and FoxO1a—in rat cardiomyocytes.51 Given the essential role of PI3K signaling in skeletal myogenesis,106 it was a reasonable prediction that miR-486 would be required therein. Indeed, miR-486 has recently been shown to positively regulate myoblast differentiation.52 However, in that study Pax7 is identified as the direct target of miR-486,52 along with miR-206 (discussed earlier).
Emerging Themes of miRNA Regulation in Myogenesis
Some common themes have emerged from our current understanding of myogenic miRNA function and regulation. It is clear that these features are universal for miRNAs that function in all aspects of biology, rather than being unique to myogenesis.
Regulatory feedbacks between miRNAs and their gene targets.
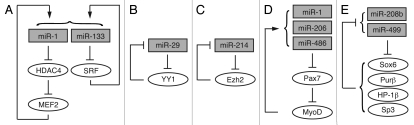
In several cases a miRNA is found to be under the regulation of its own target, forming a feedback loop. For instance, miR-1 targets HDAC4 and subsequently activates MEF2, which in turn stimulates miR-1 expression; at the same time, miR-133 targets SRF, a regulator of miR-133 transcription (Fig. 1A).40 MiR-29 expression is suppressed by its target YY1 (Fig. 1B).54 During myogenic differentiation, miR-29 is induced and it inhibits the expression of YY1, resulting in a robust upregulation of this myogenic miRNA.54,107 In rhabdomyosarcoma cells and primary tumors where YY1 is elevated, miR-29 is silenced and differentiation is therefore impaired.54,107 Similarly, miR-214 expression is suppressed in myoblasts by its target Ezh2. Upon differentiation Ezh2 is disengaged from the miR-214 promoter, which is then occupied and activated by MyoD and myogenin, leading to miR-214 expression. MiR-214 in turn downregulates Ezh2 expression, forming a feedback loop ensuring the progression of myogenic program (Fig. 1C).53 In the case of miR-1, miR-206 and miR-486 (Fig. 1D), they all target Pax7,47,52 which promotes the expression of Id2, an inhibitor of MyoD; the derepression of MyoD in turn activates the expression of these miRNAs, driving a MyoD-dependent differentiation state.52 Another example is the intronic MyomiRs, miR-208b and miR-499 (Fig. 1E), that are expressed along with their host myosin genes and target the repressors of the transcription of those genes, thus effecting myofiber type specification and muscle performance.49 This mode of feedback is likely to ensure robustness of responses that drive developmental switches such as myogenic differentiation.
Figure 1.
MiRNAs and their targets form feedback loops in the regulation of skeletal myogenesis. See text under “Regulatory feedbacks between miRNAs and their gene targets” for details.
Multiplicity of miRNA targeting.
The combinatorial effects of co-functioning miRNAs and their multi-gene targeting are one of the defining features of miRNA regulation. MiRNAs having a common seed sequence may share gene targets, which can result in redundancy or cooperation of regulation. Myogenic miRNAs characterized so far that fall into that category include (pair-wise) miR-1/miR-206, miR-221/miR-222, miR-208/miR-499 and miR-322/424/miR-503. In addition, many myogenic miRNAs have closely related isoforms that most likely share gene targets and therefore function redundantly, although not all of them have been studied. The contribution of each miRNA isoform to a specific process will likely depend on the expression level of that isoform. For instance, of the two miR-125 isoforms, miR-125b appears to play a dominant role in negatively regulating skeletal myogenesis while miR-125a levels in both myoblasts and myotubes are very low.40,66,108 The functional redundancy of miRNAs extends beyond miRNA families, as it has become apparent that a gene can be simultaneously targeted by multiple miRNAs with unrelated sequences. Examples discussed in this review include Pax7 targeting by miR-1/miR-206 and miR-486, and Ezh2 targeting by miR-24 and miR-214. Conversely, a single miRNA often exerts its function by targeting multiple genes. MiR-1 and miR-206 illustrate this principle nicely, their reported targets including a DNA polymerase, a gap junction protein, a muscle structural protein, a transcriptional regulator, an oncogenic receptor tyrosine kinase and others. It is expected that many more cases of multi-targeting in both modes will be discovered, as myogenic miRNAs continue to be characterized.
Concluding Remarks
Over the last decade, our insights into miRNA regulation and function have grown exponentially, with paradigms being established, although working models are still constantly modified by the continuous output of a tremendous volume of data. The regulatory roles of miRNA have been established in almost every aspect of skeletal muscle development,109 with a dozen or so myogenic miRNAs identified and their targets revealed (Table 1). However, this is unlikely to be a complete list. Future work, partly inspired by the sizable number of miRNAs found to be differentially expressed during skeletal myogenesis and powered by recent technological advances in biochemical purification of miRNA targets and deep sequencing,77 will likely expand the family of myogenic miRNAs significantly. As our knowledge of individual myogenic miRNAs accumulates, it will be desirable and potentially feasible, to piece together a network of miRNAs and their targets and achieve an understanding of the circuitry at a systems biology level. Towards that goal, it will also be interesting to probe sufficiency of miRNA regulation—is there a subset of miRNAs that is sufficient for governing each key step of skeletal myogenesis? Adding individual or groups of miRNAs back into miRNA-depleted (e.g., Dgcr8 knockout) myocytes or muscles could help tackle this issue. Finally, as a new class of regulators of skeletal myogenesis, miRNAs hold the potential for the development of novel biomarkers and therapeutic strategies for muscular diseases.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 6.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 7.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat Genet. 2006;38:2–7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 8.Olena AF, Patton JG. Genomic organization of microRNAs. J Cell Physiol. 2010;222:540–545. doi: 10.1002/jcp.21993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 10.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 11.Okamura K, Chung WJ, Lai EC. The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs. Cell Cycle. 2008;7:2840–2845. doi: 10.4161/cc.7.18.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle. 2010;9:4455–4460. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, et al. Diverse pathways generate microRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 20.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasudevan S, Tong Y, Steitz JA. Cell cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 25.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckingham M. Skeletal muscle formation in vertebrates. Curr Opin Genet Dev. 2001;11:440–448. doi: 10.1016/s0959-437x(00)00215-x. [DOI] [PubMed] [Google Scholar]
- 29.Weintraub H. The MyoD family and myogenesis: redundancy, networks and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 30.Naya FJ, Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol. 1999;11:683–688. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- 31.O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, et al. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res. 2009;105:585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;9:9. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. 2010;189:1157–1169. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeng SF, Rau CS, Liliang PC, Wu CJ, Lu TH, Chen YC, et al. Profiling muscle-specific microRNA expression after peripheral denervation and reinnervation in a rat model. J Neurotrauma. 2009;26:2345–2353. doi: 10.1089/neu.2009.0960. [DOI] [PubMed] [Google Scholar]
- 44.Greco S, De Simone M,, Colussi C, Zaccagnini G, Fasanaro P, Pescatori M, et al. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. Faseb J. 2009;23:3335–3346. doi: 10.1096/fj.08-128579. [DOI] [PubMed] [Google Scholar]
- 45.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol. 2007;293:451–457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 46.Yuasa K, Hagiwara Y, Ando M, Nakamura A, Takeda S, Hijikata T. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct Funct. 2008;33:163–169. doi: 10.1247/csf.08022. [DOI] [PubMed] [Google Scholar]
- 47.Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 49.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCarthy JJ, Esser KA, Peterson CA, Dupont-Versteegden EE. Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol Genomics. 2009;39:219–226. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dey BK, Gagan J, Dutta A. MiR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NFkappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G, et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 58.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 59.Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA. 2009;28:28. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by downregulation of Cdc25A. Mol Biol Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4:7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perry RL, Parker MH, Rudnicki MA. Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Mol Cell. 2001;8:291–301. doi: 10.1016/s1097-2765(01)00302-1. [DOI] [PubMed] [Google Scholar]
- 63.Ciuffini L, Castellani L, Salvati E, Galletti S, Falcone G, Alema S. Delineating v-Src downstream effector pathways in transformed myoblasts. Oncogene. 2008;27:528–539. doi: 10.1038/sj.onc.1210665. [DOI] [PubMed] [Google Scholar]
- 64.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y, Gelfond J, McManus LM, Shireman PK. Temporal MicroRNA Expression during in vitro Myogenic Progenitor Cell Proliferation and Differentiation: Regulation of Proliferation by miR-682. Physiol Genomics. 2010 doi: 10.1152/physiolgenomics.00136.2010. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Panguluri SK, Bhatnagar S, Kumar A, McCarthy JJ, Srivastava AK, Cooper NG, et al. Genomic profiling of messenger RNAs and microRNAs reveals potential mechanisms of TWEAK-induced skeletal muscle wasting in mice. PLoS ONE. 2010;5:8760. doi: 10.1371/journal.pone.0008760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 70.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 71.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC and other cell cycle genes via binding to “seedless” 3'UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38:8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 75.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 76.Ritchie W, Flamant S, Rasko JE. Predicting microRNA targets and functions: traps for the unwary. Nat Methods. 2009;6:397–398. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
- 77.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 78.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan D, Dong Xda E, Chen X, Wang L, Lu C, Wang J, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cacchiarelli D, Martone J, Girardi E, Cesana M, Incitti T, Morlando M, et al. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12:341–351. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 85.Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 86.Liu H, Chen SE, Jin B, Carson JA, Niu A, Durham W, et al. TIMP3: A physiological regulator of adult myogenesis. J Cell Sci. 2010;2010:3. doi: 10.1242/jcs.057620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X, Wang K, Chen J, Guo J, Yin Y, Cai X, et al. In vitro evidence suggests that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is an indispensable step in myogenic differentiation. J Biol Chem. 2009;284:5362–5369. doi: 10.1074/jbc.M807523200. [DOI] [PubMed] [Google Scholar]
- 89.Caretti G, Di Padova M,, Micales B,, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8:172–175. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 91.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 92.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 93.Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, et al. NFkappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol. 2007;27:4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Florini JR, Ewton DZ, Coolican SA. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- 96.Erbay E, Park IH, Nuzzi PD, Schoenherr CJ, Chen J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J Cell Biol. 2003;163:931–936. doi: 10.1083/jcb.200307158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ge Y, Wu AL, Warnes C, Liu J, Zhang C, Kawasome H, et al. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am J Physiol Cell Physiol. 2009;297:1434–1444. doi: 10.1152/ajpcell.00248.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamamoto M, Kuroiwa A. Hoxa-11 and Hoxa-13 are involved in repression of MyoD during limb muscle development. Dev Growth Differ. 2003;45:485–498. doi: 10.1111/j.1440-169x.2003.00715.x. [DOI] [PubMed] [Google Scholar]
- 101.Liu J, Luo XJ, Xiong AW, Zhang ZD, Yue S, Zhu MS, et al. MicroRNA-214 promotes myogenic differentiation by facilitating exit from mitosis via downregulation of proto-oncogene N-ras. J Biol Chem. 2010;285:26599–26607. doi: 10.1074/jbc.M110.115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2009;15:15. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 105.Messina G, Blasi C, La Rocca SA, Pompili M, Calconi A, Grossi M. p27Kip1 acts downstream of N-cadherin-mediated cell adhesion to promote myogenesis beyond cell cycle regulation. Mol Biol Cell. 2005;16:1469–1480. doi: 10.1091/mbc.E04-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang BH, Zheng JZ, Vogt PK. An essential role of phosphatidylinositol 3-kinase in myogenic differentiation. Proc Natl Acad Sci USA. 1998;95:14179–14183. doi: 10.1073/pnas.95.24.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang H, Sun H, Guttridge DC. microRNAs: novel components in a muscle gene regulatory network. Cell Cycle. 2009;8:1833–1837. doi: 10.4161/cc.8.12.8851. [DOI] [PubMed] [Google Scholar]
- 108.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 109.Crist CG, Buckingham M. microRNAs gain magnitude in muscle. Cell Cycle. 2009;8:3627–3628. doi: 10.4161/cc.8.22.9960. [DOI] [PubMed] [Google Scholar]