Abstract
The final size and shape of fruits is determined by organogenesis. Organogenesis is the coordination of cell growth, cell differentiation and pattern formation. Individual genes have been identified that affect lateral organ growth. A majority of these characterized growth genes in Arabidopsis affect all lateral plant organs and few of these have been placed into a regulatory network controlling organ growth. We have recently characterized GORDITA (GOA), a MADS-box transcription factor, which represses cell expansion specifically in fruits and affects overall fruit size.1 Here we provide insights into a possible regulatory network in which GOA can function to regulate fruit growth. We further suggest how duplicated B-sister genes; GOA and TRANSPARENT TESTA 16 (TT16) could have acquired distinct regulatory roles.
Key words: GORDITA, AGL63, TT16, MADS-box, B-sister, growth, fruit development, fruit size
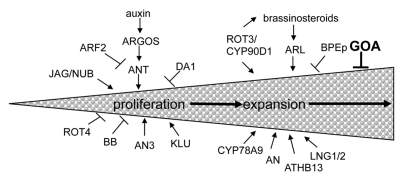
Organ morphogenesis occurs through coordinated cell growth, proliferation and cell expansion in multi-cellular organisms. Although several genes have been isolated that affect organ shape by a change in the duration of cell proliferation or a change in cell expansion (Fig. 1), there appears to be some cross-talk between cell proliferation and cell expansion by mutants that exhibit compensation.2–4 For example, an3 leaves have far fewer cells however the leaf is nearly as large as wild type because the leaf cells are larger.3–5 This compensation phenomenon suggests that there is a mechanism whereby the organ can measure its overall size and adjust its growth accordingly.4 Fruits, which are modified leaves, provide a specialized example to study the mechanisms that control lateral organ size.
Figure 1.
The size of an organ is determined by cell proliferation and cell expansion. Many proteins have been identified in Arabidopsis that play a role in these processes, however, little is known about the regulatory networks that control these processes.
A Possible Regulatory Framework of a B-Sister MADS Box Protein; GOA Action
Studies on MADS-box transcription factors have demonstrated its role in controlling organ identity not only in Arabidopsis but in many flowering plant species.6 Cell fate determining B-function MADS box transcription factors are expressed in second whorl sterile organs and third whorl male reproductive organs and they are necessary to determine the organ identity in these two whorls.7,8 Interestingly, a member of the B-sister MADS-box genes, TT16, regulates the development and pigmentation of the seed coat.9 Duplication and diversification of evolutionarily conserved transcription factors such as MADS-box proteins can bring about morphological variations among related organisms.10 Recently, we have demonstrated the functional importance of one of the duplicated B-sister MADS-box genes, GORDITA (GOA).1 We showed that the dynamic spatiotemporal expression pattern of GOA during fruit development is required to determine the final Arabidopsis fruit size largely by repressing cell expansion1 (Fig. 1). Our findings also reveal that GOA likely regulates integument differentiation additively with its paralog TT16.1,9
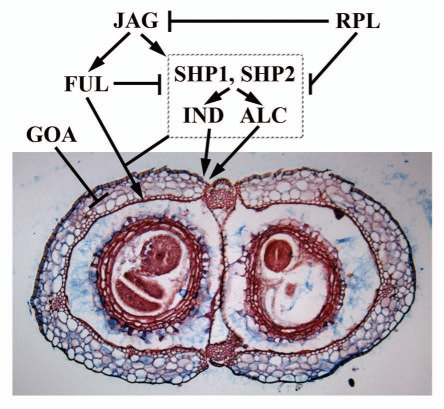
The final size and shape of an organ is likely determined by the interaction of pattern formation and growth.11 Much is known about the genes necessary for proper differentiation and pattern formation within the Arabidopsis fruit.12 FRUITFULL (FUL) is important for the proper differentiation of the valve cells and repressing the valve margin identity genes SHATTERPROOF 1 and 2 (SHP1/2)13–15 (Fig. 2). ful fruits are indehiscent and the valve cells are small and lignified due to the ectopic expression of SHP1/2. SHP1/2 are valve margin identity genes and shp1 shp2 double mutants are indehiscent due to a lack of lignification of the valve margin cells.15 INDEHISCENT (IND) is also important for valve margin identity and ind fruits are indehiscent.16 REPLUMLESS (RPL) is required for replum identity and repressing SHP1/2 in the replum.17 rpl fruits have valve margin cells that develop in place of the replum due to the ectopic expression of SHP1/2. JAG and additional regulatory proteins activate both FUL and SHP1/2.12 Further genetic interaction of these regulators with GOA should elucidate the regulatory frame work of fruit size control via coordinated cell proliferation and cell expansion (Fig. 2). Such studies should further reveal how organ size controlling mechanisms is linked to patterning and organogenesis.
Figure 2.
Patterning and growth during Arabidopsis fruit development. SHP1, SHP2, IND and ALC are necessary for patterning cells within the valve margin (vm). FUL represses the expression of these vm genes in the valves and RPL represses these vm genes in the replum. This sets up boundaries for patterning the various tissues of the fruit. JAG and other genes are necessary for proper FUL and vm gene expression. GOA represses fruit growth in the valves and FUL could repress GOA expression in the valves. Figure adapted from Dinneny et al. 2005.
Plant hormones likely act as morphogens that play a crucial role in the final size and shape of an organ.18 Several genes have been isolated that regulate cell expansion (Fig. 1). ARGOS-like (ARL) promotes cell expansion and is induced by plant hormone brassinosteroids.19 BIG PETALp (BPEp), a bHLH transcription factor, inhibits cell expansion specifically in petals.20 Although it has been suggested that the duration of cell proliferation is the main determinant of organ size and shape, several genes have been isolated that promote cell expansion and when mutated change the size and shape of lateral organs. Several molecular genetic studies in leaf growth have shown that cell expansion occurs in a polar direction. ANGUSTIFOLIA (AN) promotes polar cell expansion in the leaf width direction while ROTUNDIFOLIA3 (ROT3) and the closely related CYP90D1 promotes polar cell expansion in the leaf length direction independently of AN.21–25 ROT3 encodes a cytochrome P450 involved in brassinosteroid synthesis. LONGIFOLIA 1 and 2 (LNG1 and LNG2) belong to a small family of novel plant proteins.26 LNG1 and LNG2 are important regulators of leaf growth by polar cell expansion. Interestingly, LNG1 and LNG2 promote longitudinal cell expansion independently of ROT3 however LNG1/2 and ROT3 promote longitudinal cell expansion at the expense of transverse cell expansion (Fig. 1). CYP78A9, a member of the cytochrome P450 CYP78A family promotes fruit cell expansion in a longitudinal direction.27 ATHB13 is a homeobox leucine zipper protein that regulates leaf expansion by promoting transverse cell expansion.28 Although individual growth genes have been identified, little is known about the interaction of these growth genes and the pathways that control overall organ size and shape. Our findings of a role for GOA, a transcription factor, controlling fruit size largely by regulating cell expansion presents a starting point to investigate its function in conjunction with known cell expansion regulators.1 Intriguingly, overexpression of GOA deregulates cell expansion controlling genes such as LNG1, LNG2 and ROT3.1 Though perturbation in the level of these genes might reflect indirect regulatory action of GOA, the implication of ROT3 in brassinosteroid signaling makes it tempting to speculate that GOA controls fruit size via brassinosteroid signaling, at least in part. It will be interesting to uncover the regulatory mechanisms by which GOA controls final fruit size. Identifying upstream regulators that control the dynamic spatial and temporal distribution pattern of GOA transcripts and down stream targets of GOA protein should reveal the regulatory framework of GOA action.
B-Sister Gene Duplication and Acquisition of New Regulatory Role
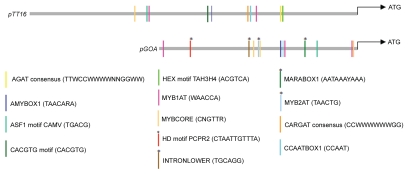
Duplicated genes can acquire new divergent functions by their ability to regulate a distinct set of downstream targets or by acquiring new domains of expression.29 TT16 and GOA are paralogs that have unique regulatory functions as reflected in their individual loss-of-function mutants, which suggest their unique targets.1,9 It is important to note that overexpression of either TT16 or GOA results in reduced plant organ growth suggesting both transcription factors should have significant overlapping regulatory targets.1,9 Isolation and further characterization of overlapping yet distinct down stream targets of TT16 and GOA should shed light on the evolutionary significance of B-sister gene duplication. Though the spatiotemporal expression pattern of TT16 is yet not known, its functional analysis suggests that some of TT16 expression domains should be distinct from that of GOA.9 In accordance we find some distinct putative cis—regulatory elements present in the promoter of GOA but not in TT16 and vice versa (Fig. 3). These predictive analyses exemplify how duplicated genes could acquire new domains of expression. Although the functional validation and importance of putative cis-elements unique to the GOA promoter still need to be characterized, the presence of potential binding sites (HD motif PCPR2) for Arabidopsis homeodomain protein PRHA and the MYB2AT motif of Persimmon DkMyb4 provide a starting point for unraveling the network involved in the GOA mediated control of fruit size.30,31 It would not be surprising if a combination of both, altered protein sequence and unique cis-regulatory sequences, are utilized by Arabidopsis B-sister duplicated paralogs to deliver new regulatory roles in addition to their conserved function.
Figure 3.
Schematic diagram displaying distribution patterns of putative cis-regulatory elements in the promoter region of TT16 (predicted promoter 3.9 kb) and GOA (1.7 kb). These analyses were carried out using PLACE (www.dna.affrc.go.jp/PLACE; Plant Cis-acting Regulatory DNA Elements). Asterisks at vertical bars represents predicted putative cis-regulatory element present only in pGOA but not in pTT16.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12095
References
- 1.Prasad K, Zhang X, Tobón E, Ambrose BA. The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. Plant J. 2010;62:203–214. doi: 10.1111/j.1365-313X.2010.04139.x. [DOI] [PubMed] [Google Scholar]
- 2.Tsukaya H. Organ shape and size: a lesson from studies of leaf morphogenesis. Curr Opin Plant Biol. 2003;6:57–62. doi: 10.1016/s1369526602000055. [DOI] [PubMed] [Google Scholar]
- 3.Tsukaya H. Leaf shape: genetic controls and environmental factors. Int J Dev Biol. 2005;49:547–555. doi: 10.1387/ijdb.041921ht. [DOI] [PubMed] [Google Scholar]
- 4.Tsukaya H. Controlling size in multicellular organs: focus on the leaf. PLoS Biol. 2008;6:174. doi: 10.1371/journal.pbio.0060174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- 6.Goto K, Kyozuka J, Bowman JL. Turning floral organs into leaves, leaves into floral organs. Curr Opin Genet Dev. 2001;11:449–456. doi: 10.1016/s0959-437x(00)00216-1. [DOI] [PubMed] [Google Scholar]
- 7.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- 9.Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, et al. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14:2463–2479. doi: 10.1105/tpc.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyerowitz EM. Pattern formation in plant development: four vignettes. Curr Opin Genet Dev. 1994;4:602–608. doi: 10.1016/0959-437x(94)90079-i. [DOI] [PubMed] [Google Scholar]
- 12.Dinneny JR, Yanofsky MF. Drawing lines and borders: how the dehiscent fruit of Arabidopsis is patterned. Bioessays. 2005;27:42–49. doi: 10.1002/bies.20165. [DOI] [PubMed] [Google Scholar]
- 13.Ferrandiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289:436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- 14.Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development. 1998;125:1509–1517. doi: 10.1242/dev.125.8.1509. [DOI] [PubMed] [Google Scholar]
- 15.Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- 16.Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Ostergaard L, Guimil S, et al. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116:843–853. doi: 10.1016/s0092-8674(04)00217-x. [DOI] [PubMed] [Google Scholar]
- 17.Roeder AH, Ferrandiz C, Yanofsky MF. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol. 2003;13:1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 18.Mizukami Y. A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol. 2001;4:533–539. doi: 10.1016/s1369-5266(00)00212-0. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Poh HM, Chua NH. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 2006;47:1–9. doi: 10.1111/j.1365-313X.2006.02750.x. [DOI] [PubMed] [Google Scholar]
- 20.Szecsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, et al. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006;25:3912–3920. doi: 10.1038/sj.emboj.7601270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, et al. CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 2005;41:710–721. doi: 10.1111/j.1365-313X.2004.02330.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, et al. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 2002;21:1267–1279. doi: 10.1093/emboj/21.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim GT, Tsukaya H, Saito Y, Uchimiya H. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:9433–9437. doi: 10.1073/pnas.96.16.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim GT, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998;12:2381–2391. doi: 10.1101/gad.12.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuge T, Tsukaya H, Uchimiya H. Two independent and polarized processes ofcell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development. 1996;122:1589–1600. doi: 10.1242/dev.122.5.1589. [DOI] [PubMed] [Google Scholar]
- 26.Lee YK, Kim GT, Kim IJ, Park J, Kwak SS, Choi G, et al. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development. 2006;133:4305–4314. doi: 10.1242/dev.02604. [DOI] [PubMed] [Google Scholar]
- 27.Ito T, Meyerowitz EM. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in arabidopsis. Plant Cell. 2000;12:1541–1550. doi: 10.1105/tpc.12.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson J, Johannesson H, Engstrom P. Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol Biol. 2001;45:247–262. doi: 10.1023/a:1006464907710. [DOI] [PubMed] [Google Scholar]
- 29.Zang J. Evolution by gene duplication. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
- 30.Plesch G, Stormann K, Torres JT, Walden R, Imre E, Somssich IE. Developmental and auxin-induced expression of the Arabidopsis prha homeobox gene. Plant J. 1997;12:635–647. doi: 10.1046/j.1365-313x.1997.00635.x. [DOI] [PubMed] [Google Scholar]
- 31.Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, et al. DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 2009;151:2028–2045. doi: 10.1104/pp.109.146985. [DOI] [PMC free article] [PubMed] [Google Scholar]