Abstract
Basic-helix-loop-helix (bHLH) proteins are a large family of eukaryotic transcription factors. In plants, they have been shown to be key regulators of a diverse array of developmental and metabolic pathways. We have recently shown that the diversity of bHLH proteins in angiosperms is ancient. Most of the bHLH subfamilies present in seed plants such as Arabidopsis thaliana and Oryza sativa are also present in early diverging groups of land plants, including mosses and lycophytes. In contrast, the diversity of bHLH proteins is much lower in chlorophytes (green algae) and red algae. This suggests that the bHLH family underwent a large expansion before or soon after the appearance of the first land plants, but has subsequently remained relatively conserved throughout the evolution of plants on land. These observations support the developing paradigm that land plants (and other complex multicellular organisms) have evolved largely through the recruitment and reorganization of ancient gene regulatory networks.
Key words: bHLH, transcription factor, plants, evolution, multicellularity, protein evolution
Basic-helix-loop-helix (bHLH) proteins are transcriptional regulators found in eukaryotic organisms. They form the second largest family of transcription factors in plants, where they are key regulators of important metabolic, physiological and developmental processes. Our recent phylogenetic study of bHLH proteins has showed how these proteins have evolved in plants.1 By analyzing bHLH coding sequences retrieved from the genomes of different algae and land plants, we were able to distinguish the main evolutionary relationships of these proteins and to infer the approximate timing of appearance of the different bHLH subfamilies.
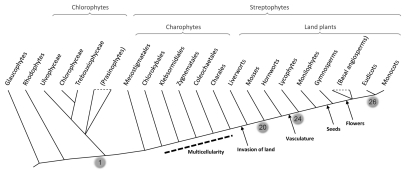
Model angiosperm species encode over 150 bHLH proteins in their genomes, which can be roughly organized in 26 smaller subfamilies. Surprisingly, we found that 20 of these bHLH subfamilies are also present in lycophytes (an ancient group of vascular plants) and mosses.1 This indicates that most of the diversity of plant bHLH was already in place in the very first land plants, before the separation of the moss and vascular plant lineages that occurred over 440 million years ago. In contrast, only a handful of bHLH sequences are encoded by the genomes of chlorophytes or red algae. This indicates that the large radiation of plant bHLH proteins occurred sometime between the separation of the chlorophyte/streptophyte lineages and the establishment of plants on land (Fig. 1).
Figure 1.
Simplified cladogram showing the phylogenetic relationships between the major groups of plants (following Lewis and McCourt8 and Qiu et al.9). The grey balloons indicate the number of modern bHLH subfamilies that were present at different nodes of plant evolution. Some of the most important events in land plant evolution are indicated.
Two different scenarios may explain our observation that a large bHLH diversity existed in early land plants. The first is that the different bHLH subfamilies evolved during the transition of plants from an aquatic to a terrestrial environment. On land, plants evolved new structures and regulatory processes that allowed them to withstand challenging new environmental conditions. bHLH proteins may have regulated the development of some of these adaptations. A second, and perhaps more attractive, scenario is that the diversification of bHLH subfamilies occurred during the evolution of multicellularity in charophyte algae, long before the invasion of the land. Charopyhtes are freshwater algae and the closest relatives to land plants (Fig. 1). The first charophytes were probably unicellular,2 but a gradual transition towards a complex multicellular body took place during charophyte evolution. Many modern bHLH proteins are involved in cellular differentiation processes (both in plants and in animals). Therefore, it is reasonable to hypothesize that the diversification of the bHLH family may have played an important regulatory role in the evolution of the increased cellular diversity that accompanied the evolution of multicellularity. Support for this hypothesis comes from the fact that the initial expansion of metazoan bHLH proteins occurred simultaneously with the evolution of multicellularity in animals.3
Interestingly, a recent phylogenetic study of plant homeobox proteins has come to similar conclusions: 14 classes of plant homeodomain proteins are present in both mosses and vascular plants, but fewer are present in unicellular chlorophytes or red algae.4 Previously, Floyd and Bowman5 had shown that many of the gene families that control angiosperm development were also present in mosses and lycophytes. Similarly, Richardt et al.6 found that there are more transcription factor families in land plants than in unicellular algae. The high degree of conservation among the gene families and subfamilies that control development during land plant evolution suggests a stimulating question: how did the biological function of each of these gene families evolve? The body plan of plants has dramatically changed since the colonization of land, with the increase in complexity of the sporophyte generation and the invention of innovative structures such as vasculature, leaves, seeds and flowers.7 Since a similar set of developmental regulators has been continually used throughout land plant history, we can conclude that the evolution of land plants occurred largely through the reusing and recycling of very ancient gene regulatory networks. Interestingly, short amino acid domains, some of which had previously been shown to be involved in protein-protein interactions, are also conserved across land plant bHLH proteins.1 This suggests that many present-day bHLH interactions also occurred in early land plants and have been conserved in the major plant groups. Since these protein-protein interactions are central to the activity of complex regulatory networks, it is reasonable to conclude that these networks have been at least partially conserved during the last 400 million years.
Today's availability of whole genome sequences from different plant species opens many doors for understanding the evolution of gene regulatory networks in land plants. Furthermore, the discovery of conservation of protein diversity and protein interactions among land plants suggests that many aspects of gene regulatory networks are conserved. Therefore, the comparative analysis of regulatory networks in a diversity of model organisms can be used to identify fundamental principles that underpin a plethora of processes in land plants. Once identified, these general principles will be critical for the sensible development of innovative technologies in economically important crop species.
Acknowledgements
This work was supported by a grant (SFRH/BD/28100/2006) to N.P. from the Fundação para a Ciência e a Tecnologia (Portugal); and grants to L.D. from the Biotechnology and Biological Research Council and Natural Environment Research Council of the United Kingdom.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12100
References
- 1.Pires N, Dolan L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol Biol Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu Y-L. Phylogeny and evolution of charophytic algae and land plants. J Syst Evol. 2008;43:287–306. [Google Scholar]
- 3.Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Curr Opin Genet Dev. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee K, Brocchieri L, Burglin TR. A Comprehensive Classification and Evolutionary Analysis of Plant Homeobox Genes. Mol Biol Evol. 2009;26:2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floyd SK, Bowman JL. The ancestral developmental tool kit of land plants. Int J Plant Sci. 2007;168:1–35. [Google Scholar]
- 6.Richardt S, Lang D, Reski R, Frank W, Rensing SA. PlanTAPDB, a Phylogeny-Based Resource of Plant Transcription-Associated Proteins. Plant Physiol. 2007;143:1452–1466. doi: 10.1104/pp.107.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langdale JA. Evolution of developmental mechanisms in plants. Curr Opin Genet Dev. 2008;18:368–373. doi: 10.1016/j.gde.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Lewis LA, McCourt RM. Green algae and the origin of land plants. Am J Bot. 2004;91:1535–1556. doi: 10.3732/ajb.91.10.1535. [DOI] [PubMed] [Google Scholar]
- 9.Qiu Y-L, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]