Abstract
The tetraploid Brassica napus possesses several seed-expressed microsomal lysophosphatidic acid acyltransferases (LPAAT) including BAT1.5, which has been retained after genome duplication as a consequence of a subfunctionalization of the gene encoding the ubiquitously expressed Kennedy pathway enzyme BAT1.13. Next, cDNA BAT1.3, encoding a LPAAT was subsequently isolated from an embryo library. The rapeseed LPAAT encoded by BAT1.3 is orthologous to the Arabidopsis thaliana At1g51260 gene product possibly associated with tapetum development and male fertility. However, BAT1.3 expression is predominant during the mid stages of embryo development in seeds of Brassica napus. Functional characterisation of BAT1.3 provides further support for a hypothesis of gene dosage sensitivity of LPAATs as does an analysis of the chromosomal localisation of LPAAT genes in Arabidopsis thaliana. The pattern of retention or loss of LPAAT genes after polyploidisation or segmental duplication is consistent with a model of balanced gene drive.
Key words: lysophosphatidic acid acyltransferase, polyploidisation, subfunctionalisation, gene loss, duplication resistant genes, balanced gene drive
Lysophosphatidic acid acyltransferase (LPAAT) performs an essential cellular function by controlling the conversion of 1-acyl-sn-glycerol-3-phosphate to phosphatidic acid, a key intermediate in the synthesis of membrane, signalling and storage lipids.1 The sub-family of plant LPAATs constitute members of a large gene family of lysophospholipid acyltransferases involved in glycerolipid synthesis and membrane lipid remodelling, although the precise function of several members of this family remains obscure.2 We are interested in the diversity and the structure and function-relationships of plant LPAATs and in their role in determining lipid acyl composition in developing seeds. In our recent manuscript,3 we have described heterogeneity among seed expressed lysophosphatidic acid acyltransferases in Brassica napus. We identified microsomal LPAAT isozymes BAT1.13 and BAT1.5 as the products of homoeologous genes and we proposed that retention of the gene pair in the tetraploid may have resulted from a subfunctionalization of one of the genes, BAT1.5 (accession no. GUO45435). The seed specific expression of BAT1.5 together with an enhanced enzymatic activity has increased the diversity of LPAAT isozymes in rapeseed and may have contributed to the capacity for lipid synthesis in this oilseed. Here we report a contrasting example of the fate of a LPAAT gene after duplication in Brassica napus.
An Unusual cDNA Encoding a Seed Expressed Microsomal LPAAT in Brassica napus
During the course of the identification of BAT1.13 and BAT1.5 we isolated a cDNA coding for an additional LPAAT from a rapeseed embryo library using a nucleotide probe corresponding to the central conserved region common to microsomal LPAATs. Remarkably, this Brassica napus cDNA is comprised of a bicistronic mRNA containing an open reading frame encoding a tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase (Bn-tRNAMeT1.3) upstream from a second ORF encoding an LPAAT, BAT1.3. The rapeseed Bn-tRNAMeT1.3 sequence is characterized by two short deletions with respect to the Arabidopsis orthologue, the first of which is predicted to result in the translation of a truncated polypeptide. Inspection of Brassica EST collections reveals that at least two distinct additional tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase genes are expressed in Brassica napus. An EST present in a seed library that is highly homologous to Bn-tRNAMeT1.3 and a second EST present in a flower bud library do not contain the frameshifting deletions present in the variant Bn-tRNAMeT1.3 sequence.
The second open reading frame in the rapeseed bicistronic cDNA codes for the BAT1.3 LPAAT which is distinct from the BAT1.13 and BAT1.5 LPAATs sharing only 58% identity and 76% similarity with these proteins. The BAT1.3 LPAAT is however, orthologous to an Arabidopsis thaliana microsomal lysophosphatidic acid acyltransferase (LPAT3)4 encoded by the At1g51260 gene sharing 89% identity and 94% similarity with this protein (UniProt accession no. Q9SYC8). The At1g51260 gene is expressed predominantly in the floral organs, during anthesis and in mature and germinating pollen and may be important for male fertility in Arabidopsis thaliana, (S. Maisonneuve and T. Roscoe, unpublished data). In contrast, the BAT1.3 LPAAT is expressed predominantly in the developing embryo in Brassica napus, raising the possibility that BAT1.3 is a subfunctionalised LPAAT and like the BAT1.5 isozyme has acquired a capacity to contribute to seed lipid synthesis.
Inactivation of a Microsomal LPAAT in Brassica napus
A detailed characterization of BAT1.3 led us to conclude that the BAT1.3 LPAAT is undergoing elimination from the genome of Brassica napus for the following reasons. That four near-identical cDNAs were isolated from two independant embryo cDNA libraries confirmed the existence of BAT1.3 as a bicistronic transcriptional unit expressed in seeds. We verified by PCR walking on Brassica napus genomic DNA that the Bn-tRNAMeT1.3 gene lies immediately adjacent and 5′ upstream from the BAT1.3 locus. In comparison, the gene At1g51260 encoding the Arabidopsis thaliana LPAT3,4 lies in the proximity of the At1g51310 gene coding for the tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase but is separated by three genes coding for hypothetical proteins on a 18 kbp region of chromosome 1. In Arabidopsis, there are many examples of adjacent loci that are transcribed to produce a chimeric pre-mRNA spanning two genes which may be spliced to generate bicistronic transcripts containing either two open reading frames or a monocistronic transcript encoding a single fusion protein.5 These unusual mRNAs may co-exist with monocistronic transcripts of the individual genes produced from such extended mRNAs. It is clear that the translation of the sequence coding for the Bn-tRNAMeT1.3 of the bicistronic transcriptional unit would result in a severely truncated protein and thus this sequence constitutes an authentic pseudogene. In contrast, translation of the BAT1.3 open reading frame would produce an intact protein of 376 residues. The BAT1.3 sequence differs from its Arabidopsis orthologue by 20 conserved and 15 non-conserved amino acid substitutions, several of which lie in residues that are invariable among LPAATs. Such variation may influence the folding, activity, substrate specificity, membrane association and orientation or protein-protein interactions of BAT1.3. The BAT1.3 protein was able to partially complement the LPAAT deficient JC201 bacterial mutant, however the activity of the recombinant BAT1.3 enzyme determined with the preferred C18:1-CoA substrate in bacterial membrane preparations was low, only slightly superior to that of control membranes. Taken together, the disrupted Bn-tMeT1.3 sequence, the possibility that BAT1.3 protein may not be translated and the near absence of enzymatic activity of recombinant BAT1.3 suggest that the bicistronic mRNA is the product of two pseudogenes. Although we cannot exclude the possibility that an additional gene, orthologous to At1g51260, preferentially expressed in anther and pollen is absent in Brassica napus, we predict that a gene encoding a LPAAT implicated in male fertility will eventually be identified.
Are Genes Encoding Lysophospholipid Acyltransferases Duplication Resistant?
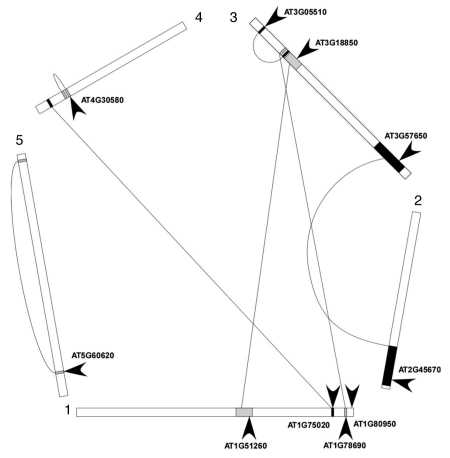
Regulation of gene dosage of LPAATs is necessary. That it occurs is confirmed by the absence of duplication of members of the lysophospholipid acyltransferase gene family in the Arabidopsis thaliana genome. The genes encoding the lysophospholipid acyltransferases are dispersed among the five chromosomes and do not show any clustering (Fig. 1). All of the genes are located in duplicated regions of the chromosomes which result either from a recent polyploidy event or from ancient segmental duplications possibly obscuring an older polyploidy.6 Eight of the genes are not duplicated in sister regions. A pair of LPAAT genes At3g57650 and At1g51260 (encoding LPAT2 and LPAT3 respectively)4 are located on segments derived from duplicated chromosomal regions where two other lysophospholipid acyltransferases At2g45670 and At3g18850 are found. Large scale sequence comparisons of their cogent BAC sequences confirmed the absence of paralogs for these genes. In each case, one of the copies has been specifically eliminated without disrupting the overall microsynteny. The gene pair At3g57650-At1g51260 are paralogs and are suggested to originate from an ancient duplication event. LPAT3,4 encoded by At1g51260 gene seems to have originated as a subfunctionalization event after duplication of At3g57650 since enzyme activity and substrate specificity are retained but the major site of expression is restricted to pollen in contrast to the ubiquitously expressed At3g57650. In summary, each gene encoding a LPAAT is unique and is not duplicated. This pattern of gene loss resembles the description of duplication resistant genes where certain genes that encode domains which when duplicated reduce fitness of the new polyploidy lineage and loss of these genes is characterized by spatially independent events.7
Figure 1.
Lysophospholipid acyltransferase genes in the Arabidopsis thaliana genome. Linked boxes depict duplicated segments overlapping LPAAT genes as defined in Blanc et al. (2003)6 (wolfe.gen.tcd.ie/athal/dup).
Conclusions
Taken together, our observations concerning evidence of gene duplication, sub-functionalization and of gene loss among LPAATs after polyploidization in Brassica napus and of elimination of LPAAT genes after duplication of chromosomal segments in Arabidopsis thaliana are consistent with the concept of dosage sensitive, ‘connected’ genes. The products of such genes are involved in protein-protein interactions and are less likely to be retained in tandem or as a transposed duplicate. More likely, they retained postpaleotetraploidy as proposed in the model of balanced gene drive.8 LPAATs are components of a tightly controlled biosynthetic pathway whose products are essential for cell viability. There is indirect evidence suggesting that LPAATs constitute components of an endomembrane located metabolon, providing intermediates for complex lipid assembly. This implies protein-protein interactions with Kennedy pathway enzymes and other lipid modifying enzymes. Such a mechanism may be invoked to explain the diversification of microsomal LPAATs and perhaps the expansion of the lysophospholipid lipid gene family.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12101
References
- 1.Hills MJ, Roscoe TJ. Synthesis of structural and storage lipids by the plant endoplasmic reticulum. In: Robinson DG, editor. Plant Cell Monographs. Vol. 4. Berlin Heidelberg: Springer-Verlag; 2006. pp. 155–186. [Google Scholar]
- 2.Roscoe TJ. Identification of acyltransferases controlling triacylglycerol biosynthesis in oilseeds using a genomics-based approach. Eur J Lipid Sci Tech. 2005;107:256–262. [Google Scholar]
- 3.Maisonneuve S, Bessoule J-J, Lessire R, Delseny M, Roscoe TJ. Expression of Brassica napus microsomal Lysophosphatidic Acid Acyltransferase isozymes enhances seed oil content in Arabidopsis thaliana. Plant Physiol. 2010;152:670–684. doi: 10.1104/pp.109.148247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim UK, Li Y, Huang AHC. Ubiquitous and endoplasmic reticulum located lysophosphatidic acid acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell. 2005;17:1073–1089. doi: 10.1105/tpc.104.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thimmapuram J, Duan H, Liu L, Schuler MA. Bicistronic and fused monocistronic transcripts are derived from adjacent loci in the Arabidopsis genome. RNA. 2005;11:128–138. doi: 10.1261/rna.7114505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 2006;22:597–602. doi: 10.1016/j.tig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]