Abstract
The plant-specific transcription factor ABSCISIC ACID IN SENSITIVE3 (ABI3) or the maize ortholog VIVIPAROUS1 (VP1) is known to regulate seed maturation and germination in concert with the phytohormone abscisic acid (ABA) but is also evolutionarily conserved among land plants including non-seed plants. An ABI3/VP1 ortholog (PpABI3A) from the moss Physcomitrella patens can activate ABA-responsive gene promoters in the moss and angiosperms; however, it failed to fully complement the phenotypes of the Arabidopsis abi3-6 mutant, suggesting that some aspects of ABI3/VP1 functions have diverged during the evolution of land plants. To gain insights into the evolution of ABI3/VP1 function, we performed a comparative analysis of the regulatory elements required for ABI3 activation in Physcomitrella using a wheat Em gene promoter, which is induced by ABA and ABI3/VP1 both in Physcomitrella and in angiosperms. Elimination of either the ACGT core motif in the ABA response element (ABRE) or the RY element, to which ABI3/VP1 binds directly, resulted in a drastic reduction of the ABA response in Physcomitrella. Arabidopsis ABI3 could effectively activate the Em promoter either in an ABRE- or RY-dependent manner, as observed in angiosperms. On the other hand, PpABI3A failed to activate an Em promoter lacking the RY element but not the ABRE. These results suggest that RY-mediated transcriptional regulation of ABI3/VP1 is evolutionarily conserved between the moss and angiosperms, whereas angiosperm ABI3/VP1 has evolved to activate ABA-inducible promoters via the ABRE sequence independently from the RY element.
Key words: ABA, ABI3, ABA response element, Physcomoitrella patens, RY element
Introduction
The plant-specific transcription factor ABSCISIC ACID INSENSITIVE3 (ABI3) or the maize ortholog VIVIPAROUS 1 (VP1) has been recognized as one of the major regulators in seed maturation, including acquisition of desiccation tolerance and dormancy.1 ABI3/VP1 integrates signaling of abscisic acid (ABA), the phytohormone indispensable for seed development, to regulate several classes of seed-specific genes, including the maturation (MAT) class and the late embryogenesis abundant (LEA) class.2,3 Although ABI3/VP1 mainly functions in seed development, ABI3 also has broader functions, such as plastid development, flowering time, and outgrowth of axillary meristems.4 Marella et al.5 previously showed that ABI3/VP1 is evolutionarily conserved in the moss Physcomitrella patens and that Physcomitrella ABI3/VP1 orthologs (PpABI3A, PpABI3B and PpABI3C) can activate a wheat ABA-responsive LEA (Em) gene promoter not only in Physcomitrella but also in barley aleurone cells. A recent accumulation of genome sequence information has revealed ancestral ABI3/VP1 genes in the genomes of the algae Chlamydomonas reinhardtii and Volvox carteri, although the functions of the gene products are not yet known.6 These findings suggest that ABI3/VP1 might emerge as a regulator of a central function in plant life that has yet to be identified, one that thereafter acquired a new function, such as regulation of seed development in angiosperms during the evolution of land plants.
ABI3/VP1 genes isolated from seed plants have revealed highly conserved domains, designated as B1, B2 and B3.7 The B1 domain is involved in physical interaction with the bZIP transcription factor ABI5 that binds to the ACGT core-containing ABA-responsive element (ABRE).8–10 The B2 domain contains a putative nuclear localization signal (NLS),11 is responsible for ABA-dependent activation through ABRE,12–15 and also facilitates interaction with ABI5.12 In addition, ABI3/VP1 transactivates gene promoters through the B3 DNA binding domain that recognizes the RY (or Sph) elements7,16 found in the 5′ regions of MAT genes, such as C1 in maize or 2S albumin genes in Arabidopsis and Brassica napus.7,14,16–18 The B3 domain of VP1 does not appear to be essential for ABA-regulated expression of the Em gene in maize,19 indicating that ABI3/VP1 can activate the downstream genes without direct binding to the promoters.
When these conserved domains of angiosperm ABI3/VP1 were compared with those of Physcomitrella ABI3s, we found that the B3 domain was highly conserved, whereas the B1 and B2 were less so.5 Complementation experiments with the Arabidopsis abi3-6 mutant with PpABI3A5 restored the green seed phenotype due to a lack of chlorophyll breakdown and expression of several seed storage protein genes that are regulated by Arabidopsis ABI3 (AtABI3). However, PpABI3A could not fully restore the ABA insensitivity in seed germination and failed to restore the expression of Em1 and Em6, which require ABI5 as the co-regulator. In line with these results, PpABI3A has shown a weak physical interaction with barley ABI5 in functional assays as well as in the yeast two-hybrid assay.5 These results indicated that some aspects of ABI3/VP1 functions are evolutionarily conserved but that others have diverged among land plants.
The wheat Em promoter is one of the best-characterized ABA-responsive and ABI3/VP1-activated promoters in angiosperms8,9,20 and is highly responsive to exogenous ABA or osmotic stress when introduced into Physcomitrella.21 A moss nuclear factor bound specifically to the ACGT core motif (Em1a) in the ABRE sequence was confirmed by DNase I footprinting, and mutations in Em1a reduced the ABA response in the moss.21 Moreover, we have shown that ABI3/VP1 from angiosperms or PpABI3A activates the Em promoter in Physcomitrella, suggesting the presence of a similar gene-regulation mechanism by ABA and ABI3 in Physcomitrella,5 although the detailed mechanism for the activation remains unknown.
To gain insights into the functional evolution of ABI3/VP1 in plants, we performed a comparative analysis in which the cis-elements for Em promoter activation by ABA and ABI3/VP1 were dissected in Physcomitrella and compared to those required for activation in angiosperms. We show that the ABRE and the RY element are critical for ABA and ABI3/VP1 activation in Physcomitrella but that there is a difference between Arabidopsis ABI3 (AtABI3) and PpABI3A in their dependency on these sequences: In contrast to AtABI3, which could activate the Em promoter in an ABRE-dependent manner even without the RY element, the RY element was necessary for activation by PpABI3A and ABRE had an additive effect on this activation.
Results
Both the ABRE and the RY element are required for ABA-dependent activation of the Em promoter in Physcomitrella.
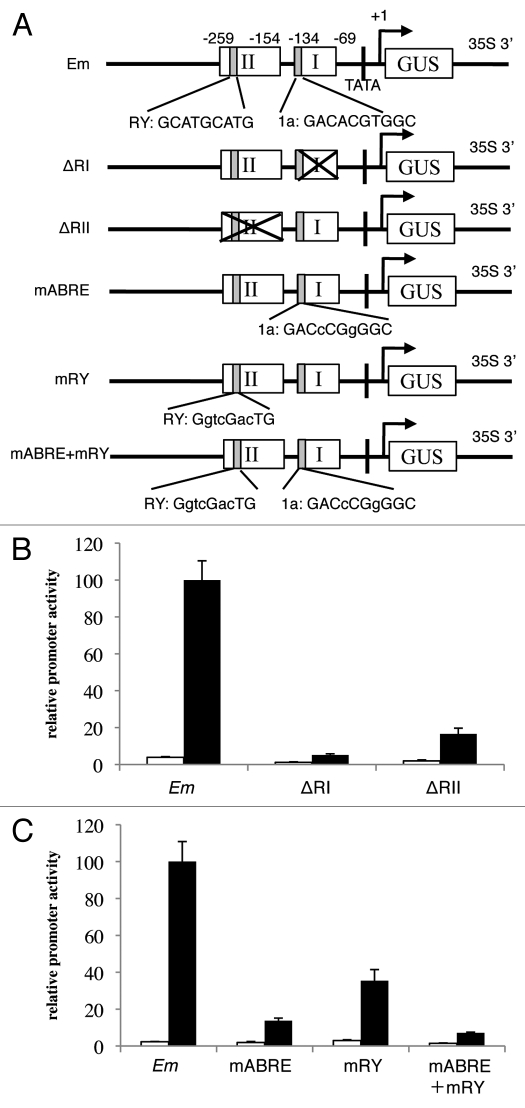
Although ABRE in the Em promoter is involved in ABA induction in Physcomitrella, the role of the RY sequence has yet to be elucidated. To determine whether ABRE is the sole determinant for ABA-dependent Em activation in Physcomitrella, we generated deletion constructs of Region I, which includes the ABRE (ΔRI), and Region II, which includes the RY element (ΔRII), respectively (Fig. 1A), and subjected them to the transient expression assay in Physcomitrella.5 Unexpectedly, both ΔRI and ΔRII failed to respond to ABA (Fig. 1B), suggesting that Region II also contains a cis-element or elements necessary for ABA induction in moss. To examine the precise cis-elements in these regions, we disrupted the Em1a motif in the ABRE and/or the RY sequence(s) by point mutations, as described.22,23 Figure 1C shows that only a 2-bp substitution in ACGT core motif in Em1a (mABRE) caused a drastic reduction in ABA inducibility, as has been shown previously;21 however, we noticed that mABRE was still significantly activated by ABA. Mutations in the RY element (mRY) also reduced ABA induction significantly in Physcomitrella, indicating that the RY element is also involved in ABA activation of the Em promoter in this species. Disruption of both sequences (mABRE + mRY) abolished most of the ABA response, suggesting that both the ABRE and the RY element are the major determinants for ABA activation of the Em promoter in Physcomitrella.
Figure 1.
Effects of deletions or mutations in the Region I and/or II of the Em promoter on activation by ABA. (A) Schematic representation of Em promoter-based reporter constructs. All promoters are fused to GUS reporter gene with a CaMV 35S terminator sequence. Sequences and the positions of the ACGT core motif in the ABRE (1a) and the RY element (RY) are shown. (B) Deletion constructs of the Region I or Region II, and (C) the substitution mutation constructs were introduced into Physcomitrella protonemal tissue by particle bombardment, then protonemal tissue was incubated with (black bars) or without (white bars) 10 µM ABA for 24 h, and the protein extracts were used for GUS and LUC assays. Bars indicate the relative GUS activities ± SE (n = 4).
Arabidopsis ABI3 activates the Em promoter in Physcomitrella in a manner similar to that in angiosperms.
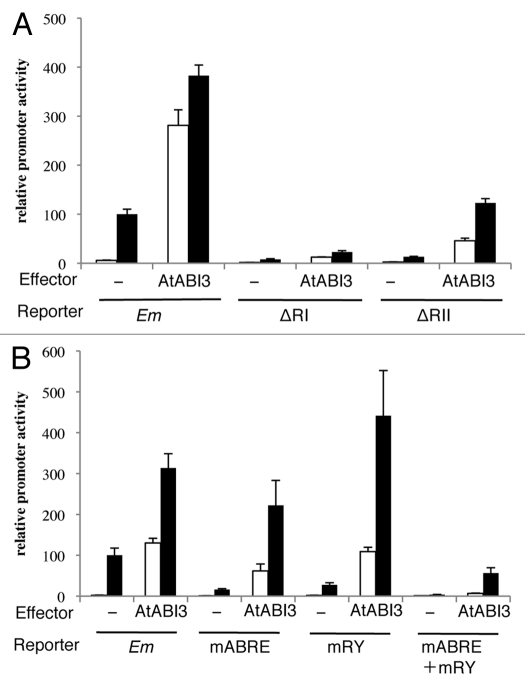
Vasil et al.9 have shown that VP1 activates the Em promoter in an ABRE-dependent manner, and Carson et al.19 have reported that the B3 domain is not essential for ABA activation of the Em gene in maize. These findings indicated that ABI3/VP1 does not require direct binding to the RY element for ABA-regulated Em gene expression in seeds. We tested the promoter activity of the ΔRI and ΔRII constructs in the presence of AtABI3 to see whether AtABI3 activates the Em promoter in an ABRE-dependent manner in Physcomitrella (Fig. 2A). Deletion of Region I greatly reduced the induction by AtABI3, but AtABI3 still significantly activated ΔRI. Deletion of Region II also affected the transactivation by AtABI3, but the reduction was less pronounced compared to that associated with the deletion of Region I.
Figure 2.
Response of Em reporter constructs to ABI3 effector. (A) Em, ΔRI and ΔRII reporter constructs, and (B) mABRE, mRY and mABRE + mRY were introduced into Physcomitrella protonemal tissue with or without the ABI3 effector construct by particle bombardment. After bombardment, the protonemal tissue was incubated with (black bars) or without (white bars) 10 µM ABA for 24 h, and the protein extracts were used for GUS and LUC assays. Bars indicate the relative promoter activity ± SE (n = 4).
These findings indicated that these two regions contain cis-acting elements for AtABI3 transactivation; however, AtABI3 can activate the Em promoter in the absence of the RY element. Because Region I and Region II contain several ACGT-core motifs, we precisely determined the involvement of Em1a in the ABRE (Region I) and/or the RY element (Region II) in the transactivation (Fig. 2B). Interestingly, mutations in either the ABRE or the RY element had no significant effect on AtABI3 transactivation; however, the simultaneous disruption of the two sequences (mABRE + mRY) greatly reduced the transactivation by AtABI3. These results suggested that AtABI3 activates the Em promoter mainly via two mechanisms in Physcomitrella; one requires the Em1a in the ABRE and the other requires the RY element.
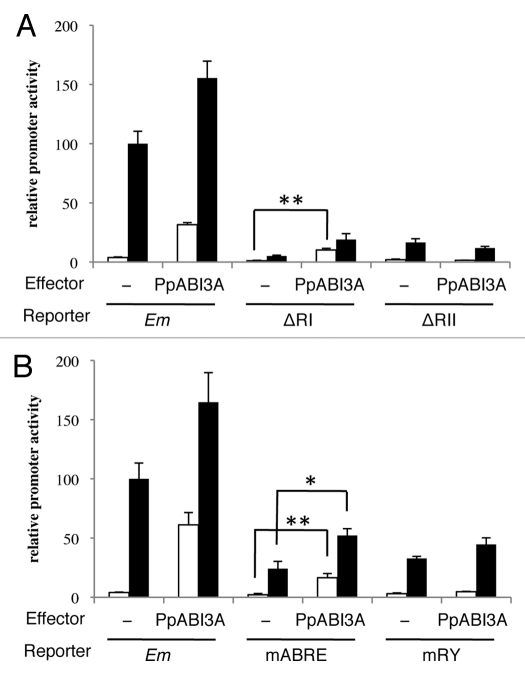
0To ascribe precisely the involvement of the ABRE and the RY element in the differential activation of the deletion constructs between PpABI3A and AtABI3, we tested the mutated promoter-GUS constructs (Fig. 3B). As expected from the experiments using the deletion constructs, PpABI3A activated the mABRE construct significantly with or without exogenous ABA, and PpABI3A barely activated the mRY construct regardless of exogenous ABA. These results suggested that, in contrast to the ABI3/VP1 regulation of the Em promoter, PpABI3A requires the RY element for ABA activation of the Em promoter in Physcomitrella, and that the ABRE is not sufficient for PpABI3A-mediated activation.
Figure 3.
Response of Em reporter constructs to PpABI3A effector. (A) Em, ΔRI and ΔRII reporter constructs, and (B) mABRE, mRY and mABRE + mRY were introduced into Physcomitrella protonemal tissue with or without the ABI3 effector construct by particle bombardment. After bombardment, the protonemal tissue was incubated with (black bars) or without (white bars) 10 µM ABA for 24 h, and the protein extracts were used for GUS and LUC assays. Bars indicate the relative promoter activity ±SE (n = 4). (*p < 0.05, **p < 0.01).
Discussion
Previous reports have shown that the wheat Em promoter is ABA-responsive in Physcomitrella and that the Em1a site in the ABRE is responsible for the induction. In addition, our detailed analysis of the promoter has revealed the RY element as another ABA-responsive element in Physcomitrella. Abolishing both the ABRE and the RY sequences drastically reduced ABA induction of the promoter, indicating that the two cis-elements are the major determinants for ABA-induced Em activation in Physcomitrella.
Involvement of the ABRE and the RY element in transcription regulation by ABI3/VP1 has been well studied in ABA-regulated genes expressed during seed maturation in angiosperms. In Arabidopsis, seed maturation can be divided into two stages; first, storage proteins (MAT) accumulate, followed by accumulation of LEA proteins that play a role in desiccation tolerance. ABA and ABI3 are deeply involved in the regulation of MAT genes and LEA genes24 through the ABRE and the RY element. Transcriptome and bioinformatic analyses have shown that the ABRE and the RY element are physically clustered in the 5′ upstream regions of genes regulated by VP1 and ABA25 that overlap with the MAT genes. Statistical analysis also demonstrated an overrepresentation of the RY element in promoters of MAT genes compared to those of LEA genes.26 These data suggest a strong association of the ABRE and RY elements in MAT gene promoters compared to LEA gene promoters. Indeed, the MAT genes in Arabidopsis, CRC and At2S3, which have RY elements in the 5′ upstream regions, require the B3 domain of AtABI3 for activation, whereas induction of AtEm1 and AtEm6, which have no RY sequence in their promoter regions, are B3 independent.25,27–29 These data indicate that AtABI3 can regulate the target genes in either an RY-dependent manner for MAT gene activation or an ABRE-dependent manner for LEA gene expression; in addition, the information suggests that AtABI3 does not need to be anchored on the promoter via the B3 domain for LEA gene regulation.
We showed that the RY-independent regulation of AtABI3 was reproduced in Physcomitrella. In contrast, we found that PpABI3A cannot activate the Em promoter in an ABRE-dependent manner and requires the RY element, indicating that PpABI3A needs to be anchored on the promoter for Em activation via the RY–B3 domain interaction to interact with a yet unknown transcription factor, possibly a bZIP protein, that binds to Em1a in the ABRE in Physcomitrella.
Different types of bZIP transcription factors recognize the ACGT-containing ABREs found in seed-specific gene promoters. OPAQUE2 (O2)-related bZIPs are responsible for the regulation of MAT genes,18 whereas ABI5-related bZIPs are responsible for the regulation of LEA genes.30 The ABRE-dependent activation of these gene promoters by ABI3 is mediated through interaction with these bZIPs. Phylogenetic analysis of plant bZIP genes has revealed that the moss genome lacks an ABI5-clade bZIP transcription factor and that it emerged after separation of bryophytes and vascular plants.31 Instead, the moss genome encodes several O2-related bZIP genes, suggesting that O2-related bZIPs may be involved in regulation of ABRE-mediated gene expression in Physcomitrella. It is likely that activation of the Em promoter by ABA and PpABI3A is controlled by a mechanism similar to that of MAT gene regulation in angiosperms, in which ABI3 bound to the RY sequence and O2-related bZIPs bound to the ABRE interact with each other. This idea agrees well with our previously reported finding that PpABI3A restored expression of O2-regulated MAT genes (CRC and At2S3) but not of ABI5-regulated LEA genes (AtEm1 and AtEm6) in the Arabidopsis abi3-6 mutant; we also identified a weak interaction of PpABI3A with ABI5-related bZIPs, which emerged after the separation of bryophytes and vascular plants. Taking these data together, we conclude that PpABI3A is functionally equivalent to Arabidopsis ABI3 in terms of B3 domain-mediated gene regulation, whereas there is a difference between these two in terms of B3 domain-independent gene regulation.
Recently Romanel et al.6 proposed that the founding member of the B3 gene family is likely similar to the ABI3/HSI (High-level expression of sugar-inducible) genes found in algae. Although the function of the ancestral ABI3s has yet to be elucidated, the recent finding that PpABI3s are essential for ABA-mediated desiccation tolerance of Physcomitrella demonstrated that ABI3/VP1 evolved to protect cells from rehydration damage after desiccation by regulating LEA-related genes in concert with ABA.32 It should be noted that ACGT core motifs are overrepresented in the 5′ upstream regions of 25 genes that are most strongly upregulated by ABA in Physcomitrella; however, the RY elements are not enriched in these promoters. Moreover, transcriptome analysis of the triple-knockout plants of the three PpABI3 genes revealed that ABA treatment significantly affected expression of fewer than 30 genes (<1% of the total number of genes examined). This number is relatively low compared to the number regulated by ectopically expressed VP1 and ABA (4.8% of the total number of genes examined).25 We speculate that the establishment of B3-independent (ABRE-dependent) transcriptional regulation would have enabled ABI3/VP1 to regulate more ABA-regulated genes in angiosperms.
In conclusion, we propose that B3 domain-mediated (RY-mediated) transcriptional regulation of ABA-regulated genes is evolutionarily conserved among land plants; in contrast, ABI3/VP1 of angiosperms has acquired B3-independent (ABRE-dependent) transcriptional regulation, possibly by acquisition of a novel interaction with newly emerged ABI5-related bZIPs that is strong enough to be independent from the RY element. It has yet to be elucidated how PpABI3s regulate their endogenous downstream genes in the genome context. Future experiments involving chromatin immunoprecipitation-sequencing will help to address this question.
Materials and Methods
Plant material.
P. patens subspecies patens (Gransden) was the wild-type strain. Protonemal tissue was grown on PpNH4 medium at 25°C under continuous light, as described previously.5
Plasmid construction.
The Em-GUS construct (p113Kp)8 with a 650-bp upstream region of the wheat Em gene fused to the β-glucuronidase (GUS) gene was used for making the promoter variants used in this study. To produce the ΔRI construct, the promoter regions from −534 to −134 and −69 to +109 were amplified by PCR using primers Em1-HindIII (5′-AAG CTT CCG CAG TTT ATT TAC GAA AAA TAG AGG G-3′) and Em4-NotI (5′-GCG GCC GCC GGC AAG GGC CTG GA-3′), and Em5-NotI (5′-GCG GCC GCG CCT CGT GCT TCA CGA-3′) and Em6-BamH1 (5′-GGA TCC CGC CAT TGC TAA CAG GTG CT-3′), respectively. The resulting promoter fragments were digested with restriction enzymes and ligated into the pBI121 vector. To generate the ΔRII construct, the promoter regions from −534 to −259 and −154 to +109 were amplified by PCR using primers Em1-HindIII (5′-AAG CTT CCG CAG TTT ATT TAC GAA AAA TAG AGG G-3′) and Em2-NotI (5′-GCG GCC GCT AGC GGC CGG GCT CG-3′), and Em3-NotI (5′-GCG GCC GCG TCT TCC AGG CCC TTG C-3′) and Em6-BamH1 (5′-GGA TCC CGC CAT TGC TAA CAG GTG CT-3′), respectively. The resulting promoter fragments were digested with restriction enzymes and ligated into the pBI121 vector.
To make the mABRE, mRY and mABRE + mRY constructs, the GeneTailor™ Site-Directed Mutagenesis System (Invitrogen, USA) was used according to the manufacturer's manual. Primers were as follows: mABRE-F (5′-CTT CCA GGC CCT TGC CGG ACC CGG GGC GCG ACA G-3′) and mABRE-R (5′-GTC CGG CAA GGG CCT GGA AGA CAA CAC GTA-3′) for mABRE; and mRY-F (5′-ACG TAC ACG CGT CGA CAA TGG TCG ACT GCA AGC AGA-3′) and mRY-R (5′-CAT TGT CGA CGC GTG TAC GTT TGT AGC GGC-3′) for mRY, respectively. The Em-GUS construct was used as the template for making mABRE and mRY, and mABRE was used as the template for the mABRE+mRY construct.
The effector constructs Act::ABI3 and Act::PpABI3A were generated as follows. The coding sequences of ABI3 and PpABI3A were amplified by PCR from the cDNAs and subcloned into the Zero Blunt TOPO vector. The amplified fragments were confirmed by sequencing, and each fragment was placed downstream of the rice Actin 1 promoter33 with the terminator of the CaMV 35S gene at the 3′ end of the fragment. The cassettes were then transferred to a vector containing the hygromycin resistance cassette.34 Ubi-LUC has been described previously.5
Transient assay.
DNA delivery to protonemal cells of P. patens was performed as described previously.5 In this study, we used 0.8 µg of each reporter construct (Em-GUS/the mutant variants and Ubi-LUC) and effector construct to prepare gold particles for four shots. One-week-old protonemal tissue was used for bombardment and then incubated on PpNH4 agar medium with or without 10 µM ABA for 24 h under the conditions described in the Plant material section. GUS and LUC activities were measured as described previously.35 GUS activity was normalized by the LUC activity and represented as relative GUS activity ±SE. All experiments consisted of four replicates.
Acknowledgements
This research was supported by Advanced Research Project of Tokyo University of Agriculture (to Y.S.).
Abbreviations
- ABA
abscisic acid
- ABI3
abscisic acid insensitive3
- ABRE
ABA-response element
- GUS
β-glucuronidase
- bZIP
basic leucine zipper
- LEA
late embryogenesis abundant
- MAT
maturation
- O2
OPAQUE2
- VP1
viviparous1
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11774
References
- 1.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes DW, Galau GA. Temporally modular gene expression during cotyledon development. Gen Dev. 1989;3:358–369. doi: 10.1101/gad.3.3.358. [DOI] [PubMed] [Google Scholar]
- 3.Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell. 1994;6:1567–1582. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohde A, Kurup S, Holdswoth M. ABI3 emerges from the seed. Trends Plant Sci. 2000;5:418–419. doi: 10.1016/s1360-1385(00)01736-2. [DOI] [PubMed] [Google Scholar]
- 5.Marella HH, Sakata Y, Quatrano RS. Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J. 2006;46:1032–1044. doi: 10.1111/j.1365-313X.2006.02764.x. [DOI] [PubMed] [Google Scholar]
- 6.Romanel EA, Schrago CG, Counago RM, Russo CA, Alves-Ferreira M. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One. 2009;4:5791. doi: 10.1371/journal.pone.0005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki M, Kao CY, McCarty DR. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcotte W, Jr, Russell SH, Quatrano RS. Abscisic acid-responsive sequences from the em gene of wheat. Plant Cell. 1989;1:969–976. doi: 10.1105/tpc.1.10.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasil V, Marcotte W, Jr, Rosenkrans L, Cocciolone SM, Vasil IK, Quatrano RS, et al. Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell. 1995;7:1511–1518. doi: 10.1105/tpc.7.9.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura S, Lynch TJ, Finkelstein RR. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001;26:627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- 11.Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill A, Nantel A, Rock CD, Quatrano RS. A conserved domain of the viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J Biol Chem. 1996;271:3366–3374. doi: 10.1074/jbc.271.7.3366. [DOI] [PubMed] [Google Scholar]
- 13.Bies-Etheve N, da Silva Conceicao A, Giraudat J, Koornneef M, Leon-Kloosterziel K, Valon C, et al. Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol Biol. 1999;40:1045–1054. doi: 10.1023/a:1006252512202. [DOI] [PubMed] [Google Scholar]
- 14.Ezcurra I, Wycliffe P, Nehlin L, Ellerstrom M, Rask L. Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 2000;24:57–66. doi: 10.1046/j.1365-313x.2000.00857.x. [DOI] [PubMed] [Google Scholar]
- 15.Marella HH, Quatrano RS. The B2 domain of VIVIPAROUS1 is bi-functional and regulates nuclear localization and transactivation. Planta. 2006;225:863–872. doi: 10.1007/s00425-006-0398-6. [DOI] [PubMed] [Google Scholar]
- 16.Dickinson CD, Evans RP, Nielsen NC. RY repeats are conserved in the 5′-flanking regions of legume seed-protein genes. Nucl Acids Res. 1988;16:371. doi: 10.1093/nar/16.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hattori T, Vasil V, Rosenkrans L, Hannah LC, McCarty DR, Vasil IK. The Viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev. 1992;6:609–618. doi: 10.1101/gad.6.4.609. [DOI] [PubMed] [Google Scholar]
- 18.Lara P, Onate-Sanchez L, Abraham Z, Ferrandiz C, Diaz I, Carbonero P, et al. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem. 2003;278:21003–21011. doi: 10.1074/jbc.M210538200. [DOI] [PubMed] [Google Scholar]
- 19.Carson CB, Hattori T, Rosenkrans L, Vasil V, Vasil IK, Peterson PA, et al. The quiescent/colorless alleles of viviparous1 show that the conserved B3 domain of VP1 is not essential for ABA-regulated gene expression in the seed. Plant J. 1997;12:1231–1240. doi: 10.1046/j.1365-313x.1997.12061231.x. [DOI] [PubMed] [Google Scholar]
- 20.Marcotte W, Jr, Guiltinan MJ, Quatrano RS. ABA-regulated gene expression: cis-acting sequences and trans-acting factors. Biochem Soc Trans. 1992;20:93–97. doi: 10.1042/bst0200093. [DOI] [PubMed] [Google Scholar]
- 21.Knight CD, Sehgal A, Atwal K, Wallace JC, Cove DJ, Coates D, et al. Molecular responses to abscisic acid and stress are conserved between moss and cereals. Plant Cell. 1995;7:499–506. doi: 10.1105/tpc.7.5.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guiltinan MJ, Marcotte W, Jr, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 23.Ezcurra I, Ellerstrom M, Wycliffe P, Stalberg K, Rask L. Interaction between composite elements in the napA promoter: both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Mol Biol. 1999;40:699–709. doi: 10.1023/a:1006206124512. [DOI] [PubMed] [Google Scholar]
- 24.Ooms J, Leon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-Insensitive abi3 mutants) Plant Physiol. 1993;102:1185–1191. doi: 10.1104/pp.102.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki M, Ketterling MG, Li QB, McCarty DR. Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 2003;132:1664–1677. doi: 10.1104/pp.103.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerriero G, Martin N, Golovko A, Sundstrom JF, Rask L, Ezcurra I. The RY/Sph element mediates transcriptional repression of maturation genes from late maturation to early seedling growth. New Phytol. 2009;184:552–565. doi: 10.1111/j.1469-8137.2009.02977.x. [DOI] [PubMed] [Google Scholar]
- 27.Kroj T, Savino G, Valon C, Giraudat J, Parcy F. Regulation of storage protein gene expression in Arabidopsis. Development. 2003;130:6065–6073. doi: 10.1242/dev.00814. [DOI] [PubMed] [Google Scholar]
- 28.Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, et al. Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 2005;46:300–311. doi: 10.1093/pcp/pci031. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, et al. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos and seedlings of Arabidopsis. Plant Mol Biol. 2006;60:51–68. doi: 10.1007/s11103-005-2418-5. [DOI] [PubMed] [Google Scholar]
- 30.Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, et al. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell. 2002;14:1391–1403. doi: 10.1105/tpc.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Correa LG, Riano-Pachon DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One. 2008;3:2944. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khandelwal K, Cho SH, Marella HH, Sakata Y, Perroud PF, Pan A, et al. Role of ABA and ABI3 in desiccation tolerance. Science. 2010;327:546. doi: 10.1126/science.1183672. [DOI] [PubMed] [Google Scholar]
- 33.McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2:163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer DG, Zryd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 35.Komatsu K, Nishikawa Y, Ohtsuka T, Taji T, Quatrano RS, Tanaka S, et al. Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol Biol. 2009;70:327–340. doi: 10.1007/s11103-009-9476-z. [DOI] [PubMed] [Google Scholar]