Abstract
Circadian clocks can be entrained by light-dark or temperature cycles. In the green alga Chlamydomonas reinhardtii, 12 h changes in temperature between 18°C and 28°C synchronize its clock. Both subunits of the circadian RNA-binding protein CHLAMY1, named C1 and C3, are able to integrate temperature information. C1 gets hyper-phosphorylated in cells grown at 18°C and the level of C3 is upregulated at this temperature. In the long period mutant per1, where temperature entrainment is disturbed, the temperature-dependent regulation of C1 and C3 is altered. Upregulation of C3 at the low temperature is mediated predominantly by an E-box element situated in its promoter region. This cis-acting element is also relevant for circadian expression of c3 as well as of its upregulation in cells, where C1 is overexpressed. Among the few identified factors interacting with the E-box region, C3 is also present, suggesting that it feedbacks on its own transcription.
Key words: Chlamydomonas reinhardtii, circadian clock, E-box element, RNA-binding protein, temperature entrainment
Introduction
Circadian rhythms occur in pro- and eukaryotic organisms. They persist with a period of about 24 h under constant conditions of light and temperature and share certain physiological properties. They can be entrained by cycles of light and darkness, and also by temperature cycles. A single pulse of e.g., light or darkness can shift their phase depending on the time the pulse is given. Moreover, circadian rhythms are temperature compensated. Thus, their period is almost unchanged at different temperatures within the physiological range of the organism under constant conditions.1
In the past years, the biflagellate alga Chlamydomonas reinhardtii emerged as a model to study the molecular mechanism of the circadian clock in a eukaryotic photosynthetic unicellular alga. The availability of its entire genome sequence allows performing studies at all levels of organization.2 Several behavioral and physiological processes are under control of an endogenous circadian clock in C. reinhardtii (reviewed in ref. 3–5). The rhythm of phototaxis that was measured with a photoaccumulation assay was one of the first described circadian rhythms in C. reinhardtii.6 The cells swim maximally to a supplied light source during subjective day, which allows them to perform optimized photosynthesis. Studies by Bruce7,8 also revealed long period mutants of the phototaxis rhythm that he named period (per). According to current knowledge, there is no connection to the well characterized PER of Drosophila melanogaster.3 Reverse and forward genetic studies revealed numerous clock-related genes in the past years, encoding several Rhythm of chloroplast (ROC) proteins,9 Constans,10 Casein kinase1,11 as well as the two subunits (named C1 and C3) of the RNA-binding protein CHLAMY1.12
Temperature Entrainment in C. reinhardtii Wild Type Cells and the per1 Mutant
Certain components of the circadian clock machinery should be able to integrate temperature information. Such a perception is needed to forward temperature information in order to entrain the circadian clock by temperature cycles, but it should be also relevant for temperature compensation. Thereby, temperature perception is needed within the physiological temperature range of an organism where the circadian clock is functioning. In some organisms, temperature changes as low as 2°C are able to communicate temperature entrainment.1 In case of C. reinhardtii, temperatures from 18°C to 28°C are still within the physiological range.13 It was shown recently that cycles of 12 h at 28°C and 12 h at 18°C (abbreviated as Thigh:Tlow) are able to entrain the biological clock of C. reinhardtii and ensure its free run under constant conditions of dim light and 18°C with a period of approx. 24 h.14 It was also found that temperature entrainment is disturbed in the long period mutant per1, resulting in a shift in acrophase under Thigh:Tlow conditions and in a low amplitude short period rhythm (approx. 22 h) under free running conditions.14
Temperature Integration of Both Subunits of CHLAMY1 and the Mechanism of c3 Upregulation
First attempts were undertaken to find out if certain components of the circadian clock can integrate temperature information. For this purpose, the expression of the two subunits, C1 and C3, of the RNA-binding protein CHLAMY1 was investigated. C1 and C3 influence phase (C3) and period (C1) of the circadian rhythms of phototaxis and nitrite reductase activity, e.g., upon their silencing.12 Interestingly, silencing or overexpression of C1 also results in parallel decrease and increase, respectively, of the C3 subunit, showing a co-regulation mechanism, while silencing or overexpression of C3 does not affect the level of C1 significantly.
To analyze potential effects of temperature on the two subunits, the cells were grown at low (18°C), medium (23°C) and high (28°C) temperatures, respectively. In case of C1, its amount is rather constant at the different temperatures, but it is phosphorylated in a temperature-dependent way. Hyper-phosphorylation occurs at 18°C and hypo-phosphorylation at 28°C.14 This phosphorylation pattern of C1 is changed in the per1 mutant where C1 does not get hyper-phosphorylated at low temperature (Table 1). In parallel, the level of C3 was analyzed in wild type cells and in the per1 mutant.14 In wild type, the C3 expression level is significantly upregulated in cells grown at 18°C in comparison to 28°C. Interestingly, the C3 expression level is increased in per1 at both temperatures in comparison to wild type, but especially at the high temperature (Table 1). These results show that both subunits of CHLAMY1 are able to integrate temperature information. They also suggest that there is a temperature-dependent functional network of the yet unknown PER1 and C1 as well as C3.
Table 1.
Temperature-dependent changes in the average degree of C1 phosphorylation (grey background) and the expression level of C3 in wild type and the per1 mutant (data adapted from Voytsekh et al.14)
| Strain | Subunit | Temperature | Percentage |
| Wild type | C1 | 18°C | 50.7 |
| Wild type | C1 | 28°C | 27.8 |
| Per1 | C1 | 18°C | 25.3 |
| Per1 | C1 | 28°C | 24.0 |
| Wild type | C3 | 18°C | 167.0 |
| Wild type | C3 | 28°C | 53.0 |
| Per1 | C3 | 18°C | 215.3 |
| Per1 | C3 | 28°C | 156.5 |
The amount of phosphorylated C1 in comparison to its non-phosphorylated form is indicated (grey background). To determine the expression level of C3 at the different temperatures in wild type and per1, the C3 level from cells grown at 23°C in wild type was set to 100% and taken for comparison.14
The mechanism of upregulation of c3 at the low temperature was under further investigation. Inhibitor experiments showed that it is regulated at the transcriptional level and this was confirmed when the c3 promoter region was put upstream to a luciferase reporter. Three elements within the c3 promoter region including its 5′ UTR were considered as potential cis-acting elements for mediating the temperature-dependent regulation (reviewed in ref. 14 and literature therein). These are two DREB1A-boxes that are recognized in higher plants by DREB transcription factors upon cold stress (4°C or below). The third one is an E-box that is known to mediate circadian regulation of the key clock components PER and Timeless in D. melanogaster. All nucleotides from the three elements were independently exchanged. Replacement of any of the DREB1A-boxes reduced the amplitude of upregulation of c3 at low temperature; however replacement of the E-box resulted in a lack of upregulation, showing that it represents the key element.
In this context, it was also checked if circadian expression of c3 that was shown by macroarray analysis to peak during subjective night phase15 is mediated by any of the three elements. Moreover, it was analyzed if the mechanism of co-regulation may involve any of these motifs. Replacement of any of the two DREB1Aboxes showed that they are not relevant for the circadian expression of c3 or for its co-regulation by the C1 subunit.16 In contrast, replacement of the E-box revealed that it represents a key element that is involved in different processes. Beside the already mentioned influence in the temperature-dependent regulation of c3, it is also necessary for its circadian expression as well as for the co-regulation in C1 overexpressing strains (Fig. 1, reviewed in ref. 14 and 16).
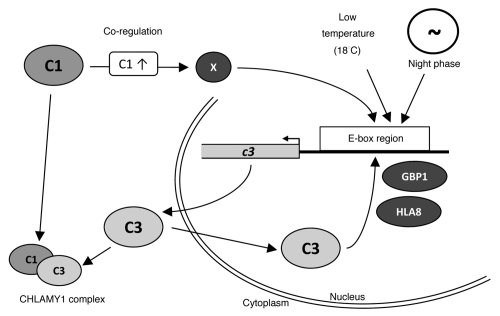
Figure 1.
Multiple roles of the E-box region. The C1 and C3 subunits of the circadian RNA-binding protein CHLAMY1 are shown and their distribution in the cytoplasm and nucleus is indicated. All factors that bind to the E-box region of the c3 promoter are depicted as well as the conditions for increasing c3 gene expression via the E-box region. These are low temperature, night phase within a circadian cycle and the co-regulation of c3 upon overexpression of C1. Since C1 was not found in nuclear extracts,16 a factor X is postulated that is involved in upregulation of c3 within the co-regulation mechanism. Additionally, the CHLAMY1 complex formation by interaction of C1 and C3 in the cytoplasm is shown. The CHLAMY1 complex binds to UG ≥7 repeat sequences of mRNAs in a circadian manner and thus influences their expression (reviewed in ref. 5).
Proteins Interacting with the E-Box Region in the c3 Promoter
Based on data of mobility shift assays along with the E-box region, an affinity approach was undertaken to identify factors that interact specifically with this region. Thereby, the E-box and the only two nucleotides distinct DREB1A-box were used as bait. Two factors were identified by mass spectrometry analysis in two independent experiments with more than one peptide. These are a G-strand telomere-binding protein1 (GBP1) and a light inducible nuclease (HLA8) that bears a TFIIB-related domain. Immunological detection further revealed that C3 itself is also able to interact with the E-box region within its own promoter, at both, day and night. Finally, a five times in tandem repeated E-box (5x-E-box) probe that lacks the neighbored DREB1A-box was used for the binding assay. Immunological detection with anti-C3 and anti-GBP1 antibodies revealed that both C3 and GBP1 are present in a shift caused by the 5x-E-box probe that is similar in mobility to the shift with the E-box region probe.16 The presence of HLA8 in this shift could not be verified by this way since there are no antibodies available so far.
The presence of a telomere-binding protein that has been characterized before to recognize the G-strand telomere sequence (TTTTAGGG)n in C. reinhardtii17 was unexpected. GBP1 bears two RNA-recognition motif (RRM) domains and was shown to be associated with RNA and single-stranded DNA.18 Several questions arise for the mechanism of c3 transcriptional activation: (1) Do telomere binding proteins have multiple functions? (2) Can RRM domain containing RNA-binding proteins such as GBP1 and C3 be also involved in transcriptional regulation? (3) Are there basic Helix-Loop-Helix (bHLH) proteins in C. reinhardtii that typically recognize E-boxes? May they have been missed by the isolation procedure? Some possible answers can be given based on comparisons with other proteins. (1) In the well studied yeast system, RAP1 is an essential structural component of yeast telomeres. RAP1 function is modulated by the precise architecture of its binding site and its surroundings. Notably, RAP1 also functions as transcriptional activator and repressor. Its binding site, including specific co-operating factors modulate its diverse functions.19 (2) RNA-binding proteins can have indeed multiple functions. This has been demonstrated, for example, for TAR DNA-binding protein 43 (TDP-43), which is a dimeric protein with two RRM domains and can bind DNA and RNA.20 It is involved in transcriptional regulation as well as in splicing, mRNA stability, transport and translation.21–25 (3) bHLH factors do occur in C. reinhardtii, but are rare. Only four have been predicted, while in Arabidopsis predictions for 160 were made.26 Such factors may have been missed in the biochemical procedure due to very low abundance, for example. A different experimental approach may help to answer this question, in future. For example, one could conduct insertional mutagenesis with the reporter gene transgenic line that is under control of the c3 promoter region. In this line, the activity of the reporter is upregulated in cells grown at the low temperature. Insertional mutants of this line that don't show this upregulation would represent bona fide members to identify genes that are involved in the temperature-dependent control of c3. However, it may also be the case that bHLH factors are not involved and that the mechanism of c3 transcriptional activation is quite different. The presence of HLA8 together with the RRM domain bearing GBP1 and C3 might indicate some kind of similar mechanism as it was found in c-myc transcriptional control.16,27,28 HLA8 that is induced by high light has a conserved nuclease domain and InterPro additionally predicts a transcription factor TFIIB-related domain (IPR000812). In case of c-myc transcription, FUSE-binding protein (FBP) interacts with a single-stranded DNA sequence that is located upstream of the c-myc promoter. This sequence has been named far upstream element (FUSE). FBP, an RNA binding protein with four K-homology domains, is stimulating the p89 helicase subunit of the transcription factor TFIIH. An FBP-interacting repressor (FIR), which binds to the FUSE element, FBP and TFIIH, plays a role as counterbalancer. FIR is a nucleic acid binding protein with two RRM domains (reviewed in ref. 28).
Another important issue concerns the feedback of C3 on its own promoter region. Since C3 is more abundant at 18°C, when its promoter is activated, it was suggested that it acts in a positive feedback loop.16 But more experimental data are needed to make final conclusions. C3 is found in eluates interacting with the 5x-E-box from cells grown at both 18°C and 28°C. Immunological detection of C3 in nuclear extracts and in the eluates indicates posttranslational modified forms. One cannot exclude that different modified forms of C3 may determine its role as a positive and negative feedback player, respectively. An overview of the complex role of the E-box region within the transcriptional activation of c3 is provided in Figure 1.
Conclusions and Perspectives
With the C1 and C3 subunits of CHLAMY1, the first components have been identified that can integrate temperature information within the physiological range of C. reinhardtii. Their temperature-dependent regulation as well as entrainment by temperature cycles are altered in the long period mutant per1, suggesting a functional network. The central cis-acting element for upregulation of c3 at low temperature is an E-box element situated in its promoter region. This element mediates also c3 circadian expression as well as its co-regulation by the C1 subunit. GBP1, HLA8 and C3 itself can interact with the E-box region and indicate a complex mechanism of transcriptional control. It will be of special interest in future, to find out how the mechanism of temperature, circadian and co-regulation control are achieved. One possibility would be that posttranslational modifications of the same factors determine their final role or that yet unknown additional factors are recruited under certain conditions.
Acknowledgements
Our work was supported by grants of the Deutsche Forschungsgemeinschaft and the Bundesministerium für Bildung und Forschung (project GoFORSYS; WP1) to M.M. K.M.M. is supported by a fellowship of the International Leibniz Research School for Microbial and Biomolecular Interactions, Jena.
Abbreviations
- bHLH
basic helix-loop-helix domain
- DREB
dehydration responsive element binding
- FBP
FUSE-binding protein
- FIR
FBP-interacting repressor
- FUSE
far upstream element
- GBP1
G-strand telomere binding protein 1
- HLA8
high light-induced nuclease 8
- PER
period
- RAP1
repressor/activator protein 1
- ROC
rhythm of chloroplast
- RRM
RNA-recognition motif
- TDP-43
TAR DNA-binding protein 43
- TFIIB
transcription factor IIB
- TFIIH
transcription factor IIH
- Thigh:Tlow
temperature cycle of 12 h at 28°C and 12 h at 18°C
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12564
References
- 1.Rensing L, Ruoff P. Temperature effect on entrainment, phase shifting and amplitude of circadian clocks and its molecular bases. Chronobiol Int. 2002;19:807–864. doi: 10.1081/cbi-120014569. [DOI] [PubMed] [Google Scholar]
- 2.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, et al. The evolution of key animal and plant functions is revealed by analysis of the Chlamydomonas genome. Science. 2007;318:245–251. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittag M, Kiaulehn S, Johnson CH. The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 2005;137:399–409. doi: 10.1104/pp.104.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuo T, Ishiura M. New insights into the circadian clock in Chlamydomonas. Int Rev Cell Molec Biol. 2010;280:281–314. doi: 10.1016/S1937-6448(10)80006-1. [DOI] [PubMed] [Google Scholar]
- 5.Schulze T, Prager K, Dathe H, Kelm J, Kieβling P, Mittag M. How the green alga Chlamydomonas reinhardtii keeps time. Protoplasma. 2010 doi: 10.1007/s00709-010-0113-0. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Bruce VG. The biological clock in Chlamydomonas reinhardtii. J Protozool. 1970;17:328–334. [Google Scholar]
- 7.Bruce VG. Mutants of the biological clock in Chlamydomonas reinhardtii. Genetics. 1972;70:537–548. doi: 10.1093/genetics/70.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce VG. Recombinants between clock mutants of Chlamydomonas reinhardtii. Genetics. 1974;77:221–230. doi: 10.1093/genetics/77.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M. A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 2008;22:918–930. doi: 10.1101/gad.1650408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F. Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol. 2009;19:359–368. doi: 10.1016/j.cub.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt M, Geβner G, Luff M, Heiland I, Wagner V, Kaminski M, et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell. 2006;18:1908–1930. doi: 10.1105/tpc.106.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iliev D, Voytsekh O, Schmidt E-M, Fiedler M, Nykytenko A, Mittag M. A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 2006;142:797–806. doi: 10.1104/pp.106.085944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris EH. The Chlamydomonas Sourcebook. San Diego: Academic Press; 1989. [Google Scholar]
- 14.Voytsekh O, Seitz SB, Iliev D, Mittag M. Both subunits of the circadian RNA-binding protein CHLAMY1 can integrate temperature information. Plant Physiol. 2008;147:2179–2193. doi: 10.1104/pp.108.118570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kucho K, Okamoto K, Tabata S, Fukuzawa H, Ishiura M. Identification of novel clock-controlled genes by cDNA macroarray analysis in Chlamydomonas reinhardtii. Plant Mol Biol. 2005;57:889–906. doi: 10.1007/s11103-005-3248-1. [DOI] [PubMed] [Google Scholar]
- 16.Seitz SB, Weisheit W, Mittag M. Multiple roles and interaction factors of an E-box element in Chlamydomonas reinhardtii. Plant Physiol. 2010;152:2243–2257. doi: 10.1104/pp.109.149195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petracek ME, Konkel LMC, Kable ML, Berman J. A Chlamydomonas protein that binds single-stranded G-strand telomere DNA. EMBO J. 1994;13:3648–3658. doi: 10.1002/j.1460-2075.1994.tb06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston SD, Lew JE, Berman J. Gbp1p, a protein with RNA recognition motifs, binds single-stranded telomeric DNA and changes its binding specificity upon dimerization. Mol Cell Biol. 1999;19:923–933. doi: 10.1128/mcb.19.1.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piña B, Fernández-Larrea J, García-Reyero N, Idrissi FZ. The different (sur)faces of Rap1p. Mol Genet Genomics. 2003;268:791–798. doi: 10.1007/s00438-002-0801-3. [DOI] [PubMed] [Google Scholar]
- 20.Kuo P-H, Doudeva LG, Wang Y-T, Shen C-KJ, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucl Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou S-HI, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abhyankar MM, Urekar C, Reddi PP. A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues—role for TDP-43 in insulator function. J Biol Chem. 2007;282:36143–36154. doi: 10.1074/jbc.M705811200. [DOI] [PubMed] [Google Scholar]
- 23.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 24.Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, et al. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Wang I-F, Wu L-S, Chang H-Y, Shen C-KJ. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 26.Riaño-Pachón DM, Corrêa LGG, Trejos-Espinos R, Mueller-Roeber B. Green transcription factors: a Chlamydomonas overview. Genetics. 2008;179:31–39. doi: 10.1534/genetics.107.086090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Im CS, Grossman A. Identification and regulation of high light induced genes in Chlamydomonas reinhardtii. Plant J. 2001;30:301–313. doi: 10.1046/j.1365-313x.2001.01287.x. [DOI] [PubMed] [Google Scholar]
- 28.Crichlow GV, Zhou H, Hsiao H-h, Frederick KB, Debrosse M, Yang Y, et al. Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition. EMBO J. 2008;27:277–289. doi: 10.1038/sj.emboj.7601936. [DOI] [PMC free article] [PubMed] [Google Scholar]