Abstract
A broad range of peroxides generated in subcellular compartments, including chloroplasts, are detoxified with peroxidases called peroxiredoxins (Prx). The Prx are ubiquitously distributed in all organisms including bacteria, fungi, animals and also in cyanobacteria and plants. Recently, the Prx have emerged as new molecules in antioxidant defense in plants. Here, the members which belong to Prx gene family in Arabidopsis and rice are been identified. Overall, the Prx members constitute a small family with 10 and 11 genes in Arabidopsis and rice respectively. The prx genes from rice are assigned to their functional groups based on homology search against Arabidopsis protein database. Deciphering the Prx functions in rice will add novel information to the mechanism of antioxidant defense in plants. Further, the Prx also forms the part of redox signaling cascade. Here, the Prx gene family has been described for rice.
Key words: antioxidant defense, chloroplast, gene family, oxidative stress, reactive oxygen species
The formation of free radicals and reactive oxygen species (ROS) occur in several enzymatic and non-enzymatic reactions during cellular metabolism. The accumulation of these reactive and deleterious intermediates is suppressed by antioxidant defense mechanism comprised of low molecular weight antioxidants and enzymes. In photosynthetic organisms, the defense against the damage from free radicals and oxidative stress is crucial. For instance, the ROS production occurs in photosystem II with generation of singlet oxygen (1O2) and hydrogen peroxide (H2O2),1,2 photosystem I from superoxide anion radicals (O2−),3 and during photorespiration with generation of H2O2.4 ROS production may exceed under environmental stress conditions like excess light, low temperature and drought.5
The antioxidant defense mechanism is activated by antioxidant metabolities and enzymes which detoxify ROS and lipid peroxides. The detoxification of ROS can occur in various cellular compartments such as chloroplasts, mitochondria, peroxisomes and cytosol.6 The enzymes like ascorbate peroxidase, catalase, glutathione peroxidase and superoxide dismutase are prominent antioxidant enzymes.6 The peroxiredoxins (Prx) emerged as new components in the antioxidant defense network of barley.7,8 Later, Prx were studied in other plants.9–14
Prx can be classified into four different functional groups, PrxQ, 1-Cys Prx, 2-Cys Prx and Type-2 Prx.15,16 They are members of the thioredoxin fold superfamily.17,18 In this study, the prx genes found in Arabidopsis and rice genomes are been identified. The Arabidopsis genome encodes 10 prx genes classified into four functional categories, 1-Cys Prx, 2-Cys Prx, PrxQ and Type-2 Prx.13 Of these, one each of 1-Cys Prx and PrxQ, two of 2-Cys Prx (2-Cys PrxA and 2-Cys PrxB) and six Type-2 Prx (PrxA–F) are identified13 (Table 1). The members of Type-2 Prx category are more in number in Arabidopsis (Table 1).
Table 1.
A list of genes encoding peroxiredoxins in Arabidopsis thaliana L.
| Locus | Annotation | Synonym | A* | B* | C* |
| AT1G48130 | 1-Cysteine peroxiredoxin 1 (ATPER1) | 1-Cys Prx | 216 | 24081.3 | 6.603 |
| AT1G60740 | Peroxiredoxin type 2 | Type-2 PrxD | 162 | 17471.9 | 5.2297 |
| AT1G65970 | Thioredoxin-dependent peroxidase 2 (TPX2) | Type-2 PrxC | 162 | 17413.9 | 5.2297 |
| AT1G65980 | Thioredoxin-dependent peroxidase 1 (TPX1) | Type-2 PrxB | 162 | 17427.8 | 4.9977 |
| AT1G65990 | Type 2 peroxiredoxin-related | Type-2 PrxA | 553 | 62653.6 | 6.4368 |
| AT3G06050 | Peroxiredoxin IIF (PRXIIF) | Type-2 PrxF | 201 | 21445.2 | 9.3905 |
| AT3G11630 | 2-Cys Peroxiredoxin A (2CPA, 2-Cys PrxA) | 2-Cys PrxA | 266 | 29091.7 | 7.5686 |
| AT3G26060 | ATPRX Q, periredoxin Q | PrxQ | 216 | 23677.8 | 10.0565 |
| AT3G52960 | Peroxiredoxin type 2 | Type-2 PrxE | 234 | 24684.0 | 9.572 |
| AT5G06290 | 2-Cysteine Peroxiredoxin B (2CPB, 2-Cys PrxB) | 2-Cys PrxB | 273 | 29779.5 | 5.414 |
A, amino acids; B, molecular weight; C, isoelectric point.
In rice (rice.plantbiology.msu.edu/), there are 11 genomic loci which encode for Prx proteins (Table 2). These loci are distributed on chromosomes 1, 2, 4, 6 and 7 (Table 2). The two Prx, peroxiredoxin-2E-1 (LOC_Os01g24740) and 2-Cys peroxiredoxin BAS1 (LOC_Os04g33970) are annotated to contain a chloroplast precursor (Table 2). The rest of Prx genes are been annotated as “peroxiredoxin, putative, expressed” (Table 2). During the study, these genes are assigned to their functional groups based on BLAST searches against Arabidopsis protein database. The Table 3 summarizes these results wherein rice LOCs and their corresponding homologs in Arabidopsis are enlisted. A nomenclature for rice prx genes is been proposed based on protein homology %identity/similarity with their Arabidopsis counterparts (Tables 1 and 3). Interestingly, a new prx gene (LOC_Os07g15670) annotated as “peroxiredoxin, putative, expressed” is identified making the tally of prx genes to eleven in rice as compared to ten in Arabidopsis (Tables 1 and 2). The BLAST search has identified its counterpart in Arabidopsis which has been annotated as “antioxidant/oxidoreductase” (AT1G21350) in the TAIR database (www.arabidopsis.org). The rice LOC_Os07g15670 and Arabidopsis AT1G21350 share protein homology %68/78 for 236 amino acids (Table 3).
Table 2.
Genes encoding peroxiredoxins in rice
| Chromosome | Locus Id | Putative function/Annotation | A* | B* | C* |
| 1 | LOC_Os01g16152 | peroxiredoxin, putative, expressed | 199 | 20873.6 | 8.2209 |
| 1 | LOC_Os01g24740 | peroxiredoxin-2E-1, chloroplast precursor, putative | 107 | 11591.5 | 6.7906 |
| 1 | LOC_Os01g48420 | peroxiredoxin, putative, expressed | 163 | 17290.8 | 5.6828 |
| 2 | LOC_Os02g09940 | peroxiredoxin, putative, expressed | 226 | 23179.5 | 6.535 |
| 2 | LOC_Os02g33450 | peroxiredoxin, putative, expressed | 262 | 28096.9 | 5.7709 |
| 4 | LOC_Os04g33970 | 2-Cys peroxiredoxin BAS1, chloroplast precursor, putative, expressed | 122 | 13410.2 | 4.3705 |
| 6 | LOC_Os06g09610 | peroxiredoxin, putative, expressed | 266 | 28926 | 10.5097 |
| 6 | LOC_Os06g42000 | peroxiredoxin, putative, expressed | 233 | 23688.3 | 9.2059 |
| 7 | LOC_Os07g15670 | peroxiredoxin, putative, expressed | 253 | 27684.6 | 9.8545 |
| 7 | LOC_Os07g44440 | peroxiredoxin, putative, expressed | 221 | 24232.6 | 5.3618 |
| 7 | LOC_Os07g44430 | peroxiredoxin, putative | 256 | 27785.3 | 6.8544 |
A, amino acids; B, molecular weight; C, isoelectric point.
Table 3.
Identification of rice homologs of peroxiredoxins in A. thaliana
| Locus Id (Os*) | Homolog (At*) | Nomenclature | Identitity/Similarity (%) | No. of aa* compared |
| LOC_Os01g16152 | AT3G06050 | Type-2 PrxF | 73/84 | 201 |
| LOC_Os01g24740 | AT1G65980 | Type-2 PrxB | 42/59 | 77 |
| LOC_Os01g48420 | AT1G65970 | Type-2 PrxC | 74/86 | 162 |
| LOC_Os02g09940 | AT1G60740 | Type-2 PrxD | 56/72 | 166 |
| LOC_Os02g33450 | AT5G06290 | 2-Cys Prx B | 74/82 | 272 |
| LOC_Os04g33970 | AT3G11630 | 2-Cys PrxA | 92/96 | 88 |
| LOC_Os06g09610 | AT3G26060 | PrxQ | 78/89 | 159 |
| LOC_Os06g42000 | AT3G52960 | Type-2 PrxE | 61/74 | 240 |
| LOC_Os07g15670 | AT1G21350 | Antioxidant | 68/78 | 236 |
| LOC_Os07g44440 | AT1G65990 | Type-2 PrxA | 27/44 | 83 |
| LOC_Os07g44430 | AT1G48130 | 1-Cys Prx | 69/83 | 221 |
Os, Oryza sativa L.; At, Arabidopsis thaliana L.; aa, amino acids.
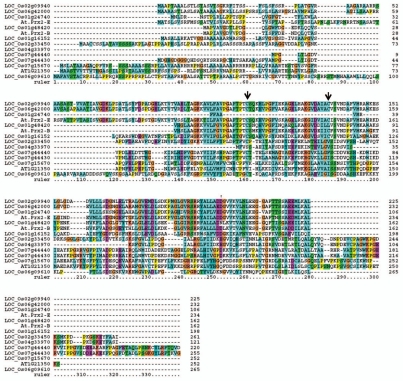
The protein alignment study of Prx members in rice with the canonical Prx2-B and Prx2-E of Arabidopsis is shown in Figure 1. The Type-2 Prx proteins are characterized by the presence of catalytic cysteine (Cys) residues (Fig. 1). The alignment of rice Prx proteins shows that the Cys residue is well conserved in members like LOC_Os02g09940 (Type-2 PrxD), LOC_Os06g42000 (Type-2 Prx E), LOC_Os01g48420 (Type-2 Prx C), LOC_Os01g16152 (Type-2 Prx F), LOC_Os02g33450 (2-Cys Prx B), LOC_Os07g44440 (Type-2 Prx A), LOC_Os07g44430 (1-Cys Prx) and LOC_Os06g09610 (PrxQ) (Fig. 1). However, LOC_Os01g24740 (Type-2 PrxB) and LOC_Os04g33970 (2-Cys PrxA) which contain a chloroplast precursor do not have the catalytic Cys residues (Fig. 1). The newly identified LOC_Os07g15670 and AT1G21350 with annotations “peroxiredoxin, putative, expressed” and “antioxidant/oxidoreductase” respectively do not have catalytic Cys residues as well (Fig. 1).
Figure 1.
Amino acid alignment of peroxiredoxins (Prx) in rice. The rice proteins are aligned with the canonical Arabidopsis Prx2-B and Prx2-E. The conserved cysteine residues are indicated by arrows on top of the alignment. Note the sequence conservation between the newly identified LOC_Os07g15670 and AT1G21350. The rice locus Ids are identified on left and amino acid positions on right. The alignment was made with ClustalX.
Taken together, the results demonstrate that like Arabidopsis, the Prx constitute a small gene family in rice. However, the functional role of Prx in rice is not clearly understood.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13494
References
- 1.Pospísil P, Arató A, Krieger-Liszkay A, Rutherford AW. Hydroxyl radical generation by photosystem II. Biochemistry. 2004;43:6783–6792. doi: 10.1021/bi036219i. [DOI] [PubMed] [Google Scholar]
- 2.Krieger-Liszkay A, Fufezan C, Trebst A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynth Res. 2008;98:551–564. doi: 10.1007/s11120-008-9349-3. [DOI] [PubMed] [Google Scholar]
- 3.Kruk J, Jemioła-Rzeminska M, Burda K, Schmid GH, Strzałka K. Scavenging of superoxide generated in photosystem I by plastoquinol and other prenyllipids in thylakoid membranes. Biochemistry. 2003;42:8501–8505. doi: 10.1021/bi034036q. [DOI] [PubMed] [Google Scholar]
- 4.Bauwe H. Recent developments in photorespiration research. Biochem Soc Trans. 2010;38:677–682. doi: 10.1042/BST0380677. [DOI] [PubMed] [Google Scholar]
- 5.Møller IM, Sweetlove LJ. ROS signaling—specificity is required. Trends Plant Sci. 2010;15:370–374. doi: 10.1016/j.tplants.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Noctor G, Foyer CH. Ascorbate and glutathione: Keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 7.Baier M, Dietz KJ. Primary structure and expression of plant homologues of animal and fungal thioredoxin-dependent peroxide reductases and bacterial alkyl hydroperoxide reductases. Plant Mol Biol. 1996;31:553–564. doi: 10.1007/BF00042228. [DOI] [PubMed] [Google Scholar]
- 8.Stacy RAP, Munthe E, Steinum T, Sharma B, Aalen RB. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol Biol. 1996;31:1205–1216. doi: 10.1007/BF00040837. [DOI] [PubMed] [Google Scholar]
- 9.Baier M, Dietz KJ. The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expression regulation, phylogenetic origin and implications for its specific physiological function in plants. Plant J. 1997;12:179–190. doi: 10.1046/j.1365-313x.1997.12010179.x. [DOI] [PubMed] [Google Scholar]
- 10.Baier M, Dietz KJ. Protective function of chloroplast 2-Cysteine peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiol. 1999;119:1407–1414. doi: 10.1104/pp.119.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee KO, Jang HH, Jung BG, Chi YH, Lee JY, Choi YO, et al. Rice 1Cys-peroxiredoxin overexpressed in transgenic tobacco does not maintain dormancy but enhances antioxidant activity. FEBS Lett. 2000;486:103–106. doi: 10.1016/s0014-5793(00)02230-4. [DOI] [PubMed] [Google Scholar]
- 12.Dietz KJ, Horling F, König J, Baier M. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J Exp Bot. 2002;53:1321–1329. [PubMed] [Google Scholar]
- 13.Dietz KJ. Plant peroxiredoxins. Annu Rev Plant Biol. 2003;54:93–107. doi: 10.1146/annurev.arplant.54.031902.134934. [DOI] [PubMed] [Google Scholar]
- 14.Rouhier N, Gelhaye E, Corbier C, Jacquot JP. Active site mutagenesis and phospholipid hydroperoxide reductase activity of poplar type II peroxiredoxin. Physiol Plant. 2004;120:57–62. doi: 10.1111/j.0031-9317.2004.0203.x. [DOI] [PubMed] [Google Scholar]
- 15.Horling F, König J, Dietz KJ. Type II peroxiredoxin C, a member of the peroxiredoxin family of Arabidopsis thaliana: Its expression and activity in comparison with other peroxiredoxins. Plant Physiol Biochem. 2002;40:491–499. [Google Scholar]
- 16.Tripathi BN, Bhatt I, Dietz KJ. Peroxiredoxins: a less studied component of hydrogen peroxide detoxification in photosynthetic organisms. Protoplasma. 2009;235:3–15. doi: 10.1007/s00709-009-0032-0. [DOI] [PubMed] [Google Scholar]
- 17.Schröder E, Ponting CP. Evidence that peroxiredoxins are novel members of the thioredoxin fold superfamily. Protein Sci. 1998;7:2465–2468. doi: 10.1002/pro.5560071125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuteja N, Umate P, Tuteja R. Conserved thioredoxin fold is present in Pisum sativum L. sieve element occlusion-1 protein. Plant Signal Behav. 2010;5:1–6. doi: 10.4161/psb.5.6.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]