Abstract
Cellulose is the most abundant biopolymer on earth. The great abundance of cellulose places it at the forefront as a primary source of biomass for renewable biofuels. However, the knowledge of how plant cells make cellulose remains very rudimentary. Cellulose microfibrils are synthesized at the plasma membrane by hexameric protein complexes, also known as cellulose synthase complexes. The only known components of cellulose synthase complexes are cellulose synthase (CESA) proteins until the recent identification of a novel component. CSI1, which encodes CESA interacting protein 1 (CSI1) in Arabidopsis. CSI1, as the first non-CESA proteins associated with cellulose synthase complexes, opens up many opportunities.
Key words: cellulose, CESA, terminal complexes, primary cell walls, Armadillo repeat
As the major load bearing polymer in cell walls, cellulose gives the plant cell its shape and maintains turgor pressure by resisting cell expansion. In growing cells, cellulose microfibrils are laid down transversely to the axis of elongation, thus forming a spring-like structure reinforcing the cell laterally and favoring longitudinal expansion.1 This orientation is crucial in determining the directionality of cell growth and ultimately plant morphology. The extent and direction of cellulose deposition is affected by many factors, including cytoskeleton, growth factors, light, mechanical stimuli, nutrition and cell-cell interactions.2–4 However, the knowledge of how these factors influence cellulose deposition remains very rudimentary. Cellulose microfibrils are thought to be synthesized at the plasma membrane by protein complexes, known as terminal complexes (TCs). The name TCs reflects the association of complexes to ends of the cellulose microfibril impression in freeze-fracture replicas. In vascular plants and some algae, TCs appears as ‘rosettes’ with a six-fold symmetry and a diameter of 25–30 nm.5–8 The first cellulose synthase (CESA) gene was discovered in A. xylinus.9,10 CESA genes have subsequently been identified in many cellulose-producing organisms including plants, algae, the slime mold Dictyostelium, bacterium and tunicates in the animal kingdom. Immuno-gold labeling of rosettes using CESA antibodies,11,12 provided convincing evidence that TCs are bona fide cellulose-synthesizing TCs. Since the first discovery of TCs in algae, much of the focus has been on the P fracture face of the plasma membrane where TCs varies in their morphology. It was only later from sectioned material that the cross-section of a linear TC in algae indicated that most of the structure was deeply embedded in the cytoplasm of the cell.13
In vascular plants, cellulose microfibrils are typically composed of approximately 36 hydrogen-bonded glucan chains. Based on the assumptions that (1) each CESA protein synthesizes one glucan chain at a time, (2) a cellulose microfibril is synthesized by a single cellulose synthase complex, (3) a single rosette (also known as CESA complex) is composed of six particles, it can be inferred that each particle is therefore occupied by six CESA proteins. However, the recent observation that cellulose synthase complexes are frequently seen as linear groupings of about four complexes that follow the same path has raised the possibility that cellulose microfibrils may be the product of a number of cellulose synthases acting coordinately. In that case, rosettes might only contain six CESA proteins rather than thirty-six. Mutant analyses in Arabidopsis have shown that a functional CESA complex has at least three types of CESAs: CESA1, CESA3 and CESA6 (or CESA6-like) are required for primary wall cellulose synthesis whereas CESA4, CESA7 and CESA8 constitute the secondary wall CESA complexes.14–17 Together with genetic evidence, biochemical studies demonstrating interactions between CESAs led to a heteromeric model of CESA complexes15,18 in which the complexes are composed of 36 individual subunits of three types of CESA isoforms. Labeling of CESA3 or CESA6 with green fluorescent protein (GFP) or yellow fluorescent protein (YFP) allows us to visualize CESA complexes in living Arabidopsis hypocotyl cells. The observation that CESA particles moved along linear trajectories at constant velocity suggest they represent actively synthesizing complexes.19
The live cell imaging of CESA complexes has led to a remarkable increase in our understanding of the enzyme's subcellular localization, regulation and trafficking during primary cell wall synthesis.19–22 It has long been argued that cortical microtubule arrays provide spatial information to the cellulose-synthesizing machinery within the plasma membrane of elongating cells.23–27 A major advance in exploring the role of microtubules in cellulose synthesis was the observation that CESA complexes can be directly observed by live-cell imaging moving through the plasma membrane upon tracks provided by the underlying cortical microtubules.19 When reorganization of microtubule arrays was triggered by exposure to light or oryzalin (a microtubule disrupting herbicide), the trajectories of the CESA particles changed accordingly. Despite the observation that alignment of cellulose microfibrils coincides with cortical microtubules, mechanistic details regarding the interactions between CESA complexes and microtubules is lacking.
The structural features of CESA proteins that are conserved from bacteria to plants include N- and C-terminal transmembrane domains and a cytoplasmic domain consisting of four conserved regions (U1 through U4) surrounding the D and QXXRW signature motif predicted to be involved in substrate binding and catalysis.28 Sequence alignments of CESAs from various organisms indicates that the plant CESAs contain three domains that are not present in any other organisms, including the N-terminal, cysteine-rich, zinc-binding domain (Zn), a conserved region (CR-P) between U1 and U2 domains, and a variable “class specific region” (CSR) between U2 and U3. The absence of CR-P and CSR region in all non-plant CESA proteins indicates that these regions are dispensable for catalytic activity of CESA proteins. Instead, these regions might mediate additional functions, e.g., interactions with other proteins in the plant cell cortex. Since a large portion of CESA proteins resides in the cytosol, rosettes are expected to be much larger on the cytosolic side than what is observed on the P fracture face of the plasma membrane. Indeed, cross sections through the plasma membrane of “bubble” algae Boergesenia forbesii revealed a very large cytoplasmic component just beneath the plasma membrane.29 We reasoned that the cytosolic region of CESA probably contained protein-protein interaction site(s) that mediated direct or indirect interactions with microtubules and regulatory proteins such as kinases that are known to act on CESA proteins. Therefore, we initiated a yeast two-hybrid screen for CESA interacting proteins using the cytosolic region of the three primary wall CESAs and identified several dozen putative CESA interacting proteins. Among them, CSI1 (CESA interacting protein 1) represents the first non-CESA protein associated with CESA complexes.30 CSI1 was initially pulled out from yeast two-hybrid screen using the central domain of CESA6. It was later confirmed that CSI1 also interacted with CESA1 and CESA3. These results suggest that CSI1 associates with CESA complexes in vivo. Indeed, CSI1 co-localizes with CESA complexes as RFP-CSI1 and GFP-CESA3 share similar localization pattern. Interestingly, RFP-CSI1 does not label any Golgi-associated CESA particles, indicating that CSI1 might not be required in the assembly of CESA complexes that presumably occurs in Golgi. RFP-CSI1 appears to be at least as abundant as GFP-CESA3, which indicate that there are more than one or two CSI1 molecules per CESA complexes. The localization pattern of RFP-CSI1 and GFP-CESA3 at the plasma membrane is not identical.30 Further detailed analysis will be needed to determine whether the difference accounts for CSI1's specific roles at the site of cellulose biosynthesis. csi1 mutants affect dynamics of CESA complexes. As shown in time-averaged projections of CESA complexes (Fig. 1), CESA complexes in csi1 do not form linear trajectories. The effect does not seem to be on the amount of complexes in the membrane but motility of complexes. Further detailed analysis will be needed to determine whether csi1 also affect the lifetime of the complexes.
Figure 1.
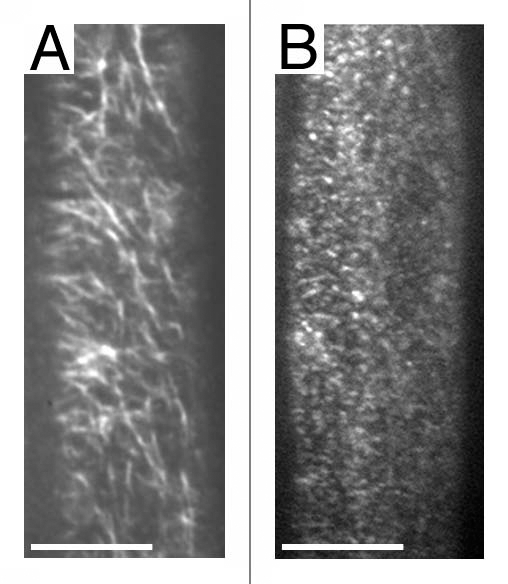
csi1 alters dynamics of CESA complexes. Time average of 61 frames (duration: 2 min; 2 s interval) showing movement of labeled particles along linear trajectories in wild type plants (A) but not in csi1-3 mutant (B). Scale bars = 5 µm.
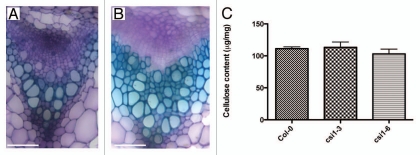
Plant cell walls are roughly classified into two types according to whether they form during cell expansion (primary cell walls) or after expansion ceases (secondary cell walls). CESA1, 3 and 6 are mainly expressed in expanding tissues whereas CESA4, 7 and 8 are expressed in xylem and vascular tissues. CSI1 is transcriptionally co-regulated with many primary CESAs including CESA1, 3, 5 and 6. However, CSI1 does not appear to be very closely co-expressed with the secondary CESA genes although CSI1 expresses also in xylem and vascular tissues. csi1-3, a T-DNA insertion null mutant of CSI1, displayed defects in cell expansion that are characterized by radially swollen epidermal cells in both primary root and dark-grown hypocotyls. Adult csi1 mutants were dwarfed as compared with wild type at/before principle growth stage 6.5.31 Defects in the secondary cell wall are characterized by a collapse of xylem vessels, a well-characterized phenotype described as irregular xylem (irx) in secondary wall cesa mutants.32 We examined cross sections of stems from both csi1 and wild type plants and we did not observe any collapsed xylem defect in csi1 (Fig. 2). Consistent with histological data, crystalline cellulose content from stems in csi1 was no different from that of wild type (Fig. 2). Thus, it appears that that csi1 has a specific role in primary cell wall biosynthesis. csi1 is viable but displays reduced fertility. There are several possibilities to explain why csi1 is not a lethal: (1) Arabidopsis encodes two CSI1-like proteins, CSI2 and CSI3. These CSI1-like proteins may be functionally redundant with CSI1. (2) CSI1 fine-tunes the orientation of cellulose deposition and is dispensable for overall cellulose biosynthesis. Additional work will be required to test these and other possibilities.
Figure 2.
csi1 has no defects associated with secondary cell wall biosynthesis. Cross sections of stems showing vascular bundles in wild type (A) and csi1-3 mutant plants (B). Stem sections were stained with toluidine blue. Bar = 50 µM. (C) Cellulose content in stems from wild type and mutant plants. Cell wall materials were collected at the late stage of stem development. Standard error bars are shown (n = 4).
CSI1 contains multiple tandem copies of a degenerate protein sequence motif, named Armadillo (ARM) repeat. The ARM repeat is an approximately 40 amino acid long tandemly repeated sequence, first identified in the Drosophila segment polarity gene armadillo.33 The Arabidopsis genome encodes more than 100 ARM repeat-containing proteins. The largest ARM repeat proteins are a subset of 41 proteins containing a N-terminal U-box motif, a motif potentially involved in proteasomal functions. PHOR1 functions in the gibberellin (GA) signaling pathway and nuclear targeting of PHOR1 requires the ARM repeat domain, whereas cytosolic localization seems to be mediated by the U-box motif.34 ARC1, another U-box containing ARM repeat protein is involved in self-compatibility (SI) signaling in Brassica. ARC1 functions in vitro as an E3 ubiquitin ligase and targets proteins for degradation during SI response.35–37 Like their animal counterparts, many plant ARM repeat protein potentially interact with the cytoskeleton. The first ARM repeat protein linked to cytoskeletal regulation in multicellular organisms was β-catenin, a component of adherens junctions in animals.38 ARABIDILLO1 and -2 are two Arabidopsis genes that show highest sequence homology to catenin. However, arabidillo-1/-2 mutant seedlings have very mild phenotype with fewer lateral roots compared with the lethal effects of catenin loss of function in animals, which leads to cell patterning defects and developmental arrest.39 ARM REPEAT ONLY1 (ARO1), is a member of a group of 28 ARM repeat proteins of Arabidopsis that appear to lack additional known protein motifs. ARO1 co-localizes with F-actin in pollen tube and may be involved in F-actin organization in tip-growing pollen tubes.40 Arabidopsis has three ARM repeat kinesin (ARK) genes, two of which are involved in epidermal cell-morphogenesis, possibly through limiting microtubule polymer levels.37
Importin-α, as the most conserved member of the family of ARM repeat proteins, is frequently used to illustrate the 3D structure of ARM repeat proteins. Three-dimensional structures of ARM repeats comprise three α helices. Importin α contains 10 ARM repeats which fold together and interact extensively with one another to form a right-handed superhelix of helices.41 Other ARM repeat-containing proteins are thought to share a conserved three-dimensional structure with importin α. By comparison, CSI1 has 10 predicted ARM repeats distributed unevenly across the entire protein (2,151 amino acids). We are not able to draw direct structural comparisons between CSI1 and other ARM repeat containing proteins. However, secondary structure prediction indicates CSI1 has extensive α helices and loops across the whole protein, which might create protein-protein interaction surfaces similar to the overall structure of other ARM repeat-containing proteins. A unique feature of the CSI1 protein is the presence of a C2 domain at the C-terminus, about 160 amino acids away from the last ARM repeat. The C2 domain is a calcium-dependent membrane-targeting module, which in many cellular proteins is involved in signal transduction or membrane targeting.42 However, C2 domains are also found in proteins that do not bind calcium, so other functions for the C2 domain, e.g., binding to inositol-1,3,4,5-tetraphosphate, have been postulated.43,44 In view of the large size of the CSI protein, we speculate that CSI1 may function as a scaffold protein that either assists in the assembly of CESA complexes, the targeting of CESA complexes to the plasmamembrane, or the interaction of the CESA complex with microtubules. Further analysis will be needed to examine the precise role of CSI1.
Acknowledgements
This work was supported in part by a grant from the US Department of Energy (DOE-FG02-03ER20133), an award from the Balzan Foundation, and startup funds from Pennsylvania State University, Department of Biochemistry & Molecular Biology.
Footnotes
Previously published online www.landesbioscience.com/journals/psb/article/13621
References
- 1.Green PB. Mechanism for plant cellular morphogenesis. Science. 1962;138:1404–1405. doi: 10.1126/science.138.3548.1404. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez-Rodriguez C, Rubio-Somoza I, Sibout R, Persson S. Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 2010;15:291–301. doi: 10.1016/j.tplants.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Seifert GJ, Blaukopf C. Irritable walls: the plant extracellular matrix and signaling. Plant Physiol. 2010;153:467–478. doi: 10.1104/pp.110.153940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derbyshire P, Findlay K, McCann MC, Roberts K. Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J Exp Bot. 2007;58:2079–2089. doi: 10.1093/jxb/erm074. [DOI] [PubMed] [Google Scholar]
- 5.Brown RM, Willison JH, Richardson CL. Cellulose biosynthesis in Acetobacter xylinum: visualization of the site of synthesis and direct measurement of the in vivo process. Proc Natl Acad Sci USA. 1976;73:4565–4569. doi: 10.1073/pnas.73.12.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giddings TH, Brower DL, Staehelin LA. Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J Cell Biol. 1980;84:327–339. doi: 10.1083/jcb.84.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller SC, Brown RM. Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J Cell Biol. 1980;84:315–326. doi: 10.1083/jcb.84.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller SC, Brown RM, Scott TK. Cellulosic microfibrils: Nascent stages of synthesis in a higher plant cell. Science. 1976;194:949–951. doi: 10.1126/science.194.4268.949. [DOI] [PubMed] [Google Scholar]
- 9.Saxena IM, Lin FC, Brown RM. Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol. 1990;15:673–683. doi: 10.1007/BF00016118. [DOI] [PubMed] [Google Scholar]
- 10.Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, et al. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 12.Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell. 1999;11:2075–2086. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudlicka K, Wardrop A, Itoh T, Brown RM. Further evidence from sectioned material in support of the existence of a linear terminal complex in cellulose synthesis. Protoplasma. 1987;136:96–103. [Google Scholar]
- 14.Gardiner JC, Taylor NG, Turner SR. Control of cellulose synthase complex localization in developing xylem. Plant Cell. 2003;15:1740–1748. doi: 10.1105/tpc.012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12:2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–780. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA. 2003;100:1450–1455. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 20.Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, et al. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21:1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, et al. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:15572–15577. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 23.Heath IB. A unified hypothesis for the role of membrane bound enzyme complexes and microtubules in plant cell wall synthesis. J Theor Biol. 1974;48:445–449. doi: 10.1016/s0022-5193(74)80011-1. [DOI] [PubMed] [Google Scholar]
- 24.Fisher DD, Cyr RJ. Extending the microtubule/microfibril paradigm. Cellulose synthesis is required for normal cortical microtubule alignment in elongating cells. Plant Physiol. 1998;116:1043–1051. doi: 10.1104/pp.116.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasteneys GO. Progress in understanding the role of microtubules in plant cells. Curr Opin Plant Biol. 2004;7:651–660. doi: 10.1016/j.pbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Herth W. Calcofluor white and Congo red inhibit chitin microfibril assembly of Poterioochromonas: evidence for a gap between polymerization and microfibril formation. J Cell Biol. 1980;87:442–450. doi: 10.1083/jcb.87.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baskin TI. On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma. 2001;215:150–171. doi: 10.1007/BF01280311. [DOI] [PubMed] [Google Scholar]
- 28.Delmer DP. CELLULOSE BIOSYNTHESIS: Exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 29.Saxena IM, Brown RM. Cellulose biosynthesis: current views and evolving concepts. Ann Bot. 2005;96:9–21. doi: 10.1093/aob/mci155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, et al. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA. 2010;107:12866–12871. doi: 10.1073/pnas.1007092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyes DC, Zayer AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, et al. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 34.Amador V, Monte E, Garcia-Martinez JL, Prat S. Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell. 2001;106:343–354. doi: 10.1016/s0092-8674(01)00445-7. [DOI] [PubMed] [Google Scholar]
- 35.Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA. 1998;95:382–387. doi: 10.1073/pnas.95.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stone SL, Arnoldo M, Goring DR. A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science. 1999;286:1729–1731. doi: 10.1126/science.286.5445.1729. [DOI] [PubMed] [Google Scholar]
- 37.Stone SL, Anderson EM, Mullen RT, Goring DR. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell. 2003;15:885–898. doi: 10.1105/tpc.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coates JC, Laplaze L, Haseloff J. Armadillo-related proteins promote lateral root development in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:1621–1626. doi: 10.1073/pnas.0507575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gebert M, Dresselhaus T, Sprunck S. F-actin organization and pollen tube tip growth in Arabidopsis are dependent on the gametophyte-specific Armadillo repeat protein ARO1. Plant Cell. 2008;20:2798–2814. doi: 10.1105/tpc.108.061028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coates JC. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 2003;13:463–471. doi: 10.1016/s0962-8924(03)00167-3. [DOI] [PubMed] [Google Scholar]
- 42.Ochoa WF, Garcia-Garcia J, Fita I, Corbalan-Garcia S, Verdaguer N, Gomez-Fernandez JC. Structure of the C2 domain from novel protein kinase Cepsilon. A membrane binding model for Ca(2+)-independent C2 domains. J Mol Biol. 2001;311:837–849. doi: 10.1006/jmbi.2001.4910. [DOI] [PubMed] [Google Scholar]
- 43.Davletov BA, Sudhof TC. A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- 44.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]