Abstract
In order for plant cells to function efficiently under different environmental conditions, chloroplastic processes have to be tightly regulated by the nucleus. It is widely believed that there is inter-organelle communication from the chloroplast to the nucleus, called retrograde signaling. Although some pathways of communication have been identified, the actual signals that move between the two cellular compartments are largely unknown. This review provides an overview of retrograde signaling including its importance to the cell, candidate signals, recent advances and current experimental systems. In addition, we highlight the potential of using drought stress as a model for studying retrograde signaling.
Key words: retrograde, chloroplast, signals, drought, stress, high light, abiotic, excess light, photosynthesis
Introduction
Compartmentalization of the eukaryotic cell into different organelles presents new paradigms in regulation of cellular function compared to prokaryotes, where all the main cellular processes happen within the same compartment. This is especially true in plant cells where chloroplasts are present in addition to mitochondria, Golgi, nucleus, ER and peroxisomes found in both plant and animal cells. In the chloroplast, many important cellular processes take place including photosynthesis, aspects of lipid, amino acid,1 chlorophyll and carotenoid biosynthesis.2 Intriguingly, however, only about 100 genes encoding for components of the chloroplast are present in the chloroplast genome; the vast majority are now nuclear-encoded.3
This compartmentalization between gene and gene product has led to the general agreement that cross-talk between the chloroplast and the nucleus must occur to co-ordinate sub cellular processes. Despite considerable research, to date no retrograde signal has been conclusively identified. This review is intended to be an introduction to retrograde signaling for readers who are new to this field. Readers interested in more detailed reviews are referred to reference 4–12. Here, we discuss the rationale for chloroplast-to-nucleus retrograde signaling, advances (and false dawns) in identifying retrograde signals and current experimental systems used in the study of retrograde signaling. This review will also examine the use of drought stress, in conjunction with mutagenesis, as a viable strategy to identify retrograde signals.
Chloroplast-to-nucleus Retrograde Signaling
Chloroplasts are endosymbiotic descendants of photosynthetic cyanobacteria-like prokaryotes that were incorporated into cells more than a billion years ago.13 Over evolutionary time, transfer of genes from the chloroplast to the nucleus has taken place,14 and today the majority of proteins functioning in the chloroplast are encoded by nuclear genes.3 These genes include those coding for components of photosynthesis,4 the sulfate assimilation pathway,15 aromatic amino acid biosynthesis,1 carotenoid biosynthesis,2 and fatty acid (FA) biosynthesis.16 Apart from these processes, the majority of the genes involved in photosynthesis and chloroplast biogenesis are also nuclear-encoded.17
The above processes in the chloroplast are all vital to the viability of the plant cell. Photosynthesis converts electrochemical energy from light into sugars and acts as the carbon source of the cell. The sulfate assimilation pathway provides the amino acid cysteine and the electron donor/acceptor, glutathione, that facilitates aspects of photosynthesis.15 Carotenoids are essential for photosynthesis and also provides the precursors to plant hormones such as strigolactones which regulate growth and shoot branching18 and abscisic acid (ABA),19 a key regulator of abiotic stress response20 and seed germination.21 FA can be modified to form integral components of cell and organellar membranes.22
Given the importance of the chloroplast to the plant, tight coupling between nuclear gene expression and chloroplast-localized processes is required. This regulation would necessitate chloroplast-nucleus communication, or ‘retrograde signaling’. It is perhaps instructive to consider this regulation in terms of different developmental stages of the chloroplast in the cell. In the simplest categorization, the stages would be immature and mature chloroplasts.
Progression from immature to mature chloroplasts, i.e., chloroplast biogenesis, represents the first stage at which chloroplast-nucleus communication is essential. Assembly of a complete, functional chloroplast during chloroplast biogenesis involves numerous nuclear-encoded processes including protein import into the chloroplast,23 assembly of thylakoid complexes complete with photosynthetic apparatus,24 accumulation of chlorophyll25 and signaling involving photoreceptors.26 Coordination of this assembly is crucial to ensure the plant cell does not waste valuable resources, for example polypeptides which are energy-intensive to synthesize. Therefore, the incomplete chloroplast and the nucleus have to communicate to build a complete chloroplast, with the plastid providing instructions on gene regulation to the nucleus. This retrograde signaling process is termed ‘biogenic control’.4
In mature chloroplasts, the need for the chloroplast to communicate with the nucleus becomes even more apparent. In essence, the chloroplast and nucleus need to communicate to keep the chloroplast functioning at optimal levels. That is, photosynthesis and other metabolic processes have to be coordinated according to fluxes in metabolites and changes in environmental conditions. This chloroplast-to-nucleus communication, and the subsequent changes in nuclear gene expression required to keep metabolic processes running “optimally” in mature chloroplasts is called ‘operational control’.4
For example, nuclear-encoded photosynthetic genes are systematically down or upregulated in response to changes in the redox status of the chloroplast.27 Similarly, inhibition of photosynthesis by application of the herbicide norflurazon results in downregulation of nuclear-encoded genes.28 Perturbation of chloroplast-localized sulfate assimilation29 via mutagenesis was also reported to result in global changes in nuclear gene expression. In the case of FA biosynthesis, de novo FA biosynthesis in the chloroplast produces acyl chains which are then transported into different cellular compartments; this flux of acyl chains is tightly regulated by nuclear genes to match the rate of FA biosynthesis.30
In addition to the vital role that retrograde signaling plays in regulating chloroplastic processes, it is also apparent that it plays a significant role in the plant's adaptive response to stresses.31 The chloroplast, by virtue of being a hub of metabolic processes, many of which are energy-intensive, is uniquely placed to act as an environmental sensor to perceive stress and coordinate the nuclear-encoded adaptive stress response. Numerous studies have investigated the role of retrograde signaling in light-stress response.32 By contrast, the role of retrograde signaling in drought stress has received scarce attention and presents fertile ground for interrogating the mechanisms of retrograde signal transduction.
What is unclear in biogenic, operational and stress-responsive retrograde signaling, however, is how such dynamic chloroplast-nucleus communication is achieved. How does the nucleus ‘sense’ what is happening in the chloroplast, and then direct the appropriate changes in protein synthesis, turnover, post-translational modifications and trafficking? A logical hypothesis would be that certain molecules that can relay information from the chloroplast to the nucleus exist, and depending on the conditions in the chloroplast such molecules may be sent from the chloroplast into the nucleus to direct changes in gene expression. Such molecules are termed ‘retrograde signals’.
Retrograde Signals: Elusive Molecules
Despite recent advances in our understanding of retrograde signaling components, actual retrograde signals have yet to be identified and there are differing views in the literature (reviewed in ref. 4–12 and 87). One of the first candidates for a retrograde signal was Mg-protoporphyrin IX (Mg-protoPIX), a chlorophyll biosynthetic intermediate. Initially, Mg-protoPIX was suggested to be involved in both biogenic and operational control. In seedlings, the perturbation of chloroplast differentiation after treatment with the herbicide norflurazon was initially reported to result in high accumulation of Mg-protoPIX.33 A similar effect in both the chloroplast and the cytoplasm was also observed during abiotic stress.34 In addition, Mg-protoPIX was thought to interact with gene products of the GENOMES UNCOUPLED (GUN) loci, which have been implicated as regulators of retrograde signaling.35,36 gun mutants continue to transcribe nuclear-encoded photosynthetic genes even when photosynthesis is inhibited by herbicide treatment, implying that retrograde signaling is impaired.37 Taken together, the Mg-protoPIX-GUN signaling cascade represented an attractive model for retrograde signaling. This theory, however, has since been challenged by Moulin et al.38 who showed that the Mg-protoPIX accumulation observed in previous studies failed to distinguish between Mg-protoPIX and its precursors. In the same year Mochizuki et al.39 also provided evidence that the steady-state level of Mg-protoPIX does not control retrograde signaling. Therefore, despite much promise, Mg-protoPIX is unlikely to be a retrograde signal, although the gun mutants have and will continue to provide insight into the process.
Another class of molecules that carry considerable interest as retrograde signal candidates, particularly in operational control, are reactive oxygen species (ROS). For a comprehensive review on ROS production, scavenging and signaling, see reference 40. In the plant cell ROS are produced from the partial reduction of atmospheric triplet oxygen (O2). There are four common forms of ROS generated by photosynthesis; these are singlet oxygen (1O2), the superoxide anion (O2·−), hydrogen peroxide (H2O2) and the hydroxyl radical (HO−·).40 Metabolically active plant organelles such as chloroplasts, peroxisomes and mitochondria generate ROS under normal conditions. In the chloroplast, during photosynthesis ROS can be generated when O2, rather than NADP+, accepts either energy or an electron from the electron transport chain.41 At photosystem (PS) II excess excitation can result in the production of singlet oxygen from triplet chlorophyll and at PSI the production of O2·- and H2O2. The extremely high reactivity of ROS enables these molecules to cause oxidative damage to multiple cellular components including proteins, lipids and DNA.42 As such, under normal conditions the plant maintains ROS to relatively low levels using a suite of ROS-scavenging proteins such as Ascorbate Peroxidase and antioxidants such as carotenoids.43 Nonetheless, the long-standing connotations of “oxidative-damage” associated with ROS are increasingly juxtaposed by the realization that ROS are essential for the well-being of organisms, including humans.44
What is interesting in the context of retrograde signaling, are the changes in nuclear gene expression that result from fluctuations in specific ROS concentrations. Different abiotic stresses ranging from drought and excess light (EL) to nutrient deprivation all decrease the maximum photosynthetic capacity of the chloroplast,32 which enhances ROS production.41,45 During abiotic stress, impaired NADP+ regeneration through the Calvin cycle causes an over reduction of the photosynthetic electron transport (PET) chain, resulting in higher leakage of electrons to O2 and consequently more ROS.
Among the four ROS species, H2O2 has probably been the best studied. This molecule has been implicated in modulating nuclear gene expression, possibly by inducing protein phosphorylation by mitogen-activated protein kinases (MAPKs),46 which are involved in signaling pathways regulating gene expression.47 Additionally, H2O2 mediates ABA signaling in response to drought stress.48,49 H2O2 is sensed by Arabidopsis glutathione peroxidase 3 (ATGPX3), which in turn modulates activities of phosphatases (e.g., ABI1), protein kinases (OST1), transcription factors and ion channels involved in ABA signaling pathways.49 In agreement with this, transcriptomic analyses of Arabidopsis plants have revealed hundreds of H2O2-responsive genes.50,51
Other ROS species are also involved in the generation of retrograde signals. Accumulation of 1O2 in the Arabidopsis fluorescent (flu) mutant results in altered expression of 70 nuclear genes.52 FLU is a negative regulator of tetrapyrrole metabolism; the mutant over accumulates the photosensitizer protochlorophyllide in the dark and consequently generates 1O2 once illuminated.53 Light-exposed flu plants exhibit induction of programmed cell death in leaves; however this is suppressed in the double mutant of flu crossed with executer 1 (ex1).54 The ex1 flu double mutant still over accumulates protochlorophyllide in the dark and releases 1O2 upon illumination, but no longer undergoes cell death.55 Taken together, these observations implicate EX1, and more importantly 1O2, in mediating a retrograde pathway that amongst other functions can regulate programmed cell death in plants.55
Gene expression arrays56 and mutations in chloroplastic copper-zinc superoxide dismutase (CuZn-SOD)57 have also suggested a role for O2·− in retrograde signaling. The specific generation of O2·− in the absence of H2O2 accumulation revealed a subset of nuclear encoded genes that are likely to be specific to an O2·− signaling pathway.56 In CuZn-SOD mutants, O2·− accumulation results in activation of chloroplast-encoded genes not stimulated by other ROS species.57 Concomitant with this is an upregulation in nuclear-encoded anthocyanin biosynthesis genes.58 These findings suggest a possible role for O2·− in mediating some aspects of retrograde signaling as well.
Could any of the ROS molecules actually be the elusive retrograde signal? An important property for a retrograde signal would be the ability to move between subcellular compartments. In this regard, among the four ROS species H2O2 would be the most ideal candidate as it has high stability and can move between cellular compartments.40 Numerous reports point towards the involvement of this molecule in inducing changes in nuclear gene expression. Furthermore, accumulation of H2O2 specifically in the chloroplast has been shown to induce expression of the nuclear-encoded genes, such as cytoplasmic APX2.59 On the other hand, to date there is no conclusive evidence to show that H2O2 acts directly on the key components of gene transcription and translation. These components include RNA polymerases, RNA processing enzymes such as exoribonucleases and ribosomes. As such, H2O2, while being an important player in signaling cascades and in generating retrograde signals, by itself is probably not a retrograde signal in the strictest sense.
Current Experimental Systems for Studying Retrograde Signaling
Chemical treatment.
The use of chemicals to inhibit particular processes specifically in the chloroplast to initiate retrograde signaling in both biogenic and operational control has been reasonably successful so far. Inhibition of chloroplastic gene expression by tagetoxin, which specifically inhibits plastid RNA polymerase, results in suppression of several nuclear genes induced during normal development, including that coding for the small subunit of Rubisco.60 A similar effect was observed when plastid translation was inhibited using lincomycin.61 For operational control, insights have been gained from the use of norflurazon,28 which inhibits chloroplastic processes and lead to the repression of nuclear-encoded photosynthetic genes in Arabidopsis. Norflurazon treatment results in inhibition of phytoene desaturase, a key enzyme in carotenoid biosynthesis.62 As carotenoids are ROS scavengers and structural components of the photosystems,43 inhibition of carotenoid biosynthesis would lead to oxidative destruction of the chloroplast.62 Additionally, the loss of carotenoid precursors for ABA biosynthesis19 would perturb signaling involving this hormone. The expectation is that such dramatic alterations of chloroplast function would lead to accumulation of retrograde signals. As discussed previously, Mg-protoPIX was first identified as a potential retrograde signal through such studies.
Perturbation of the redox state of the chloroplast via chemical treatment has also yielded useful results. The redox state of the photosynthetic electron transport chain in the chloroplast can influence nuclear gene expression.27 In particular, the redox state of the plastoquinone (PQ) pool, which mediates electron transfer between photosystems, is a critical regulator of retrograde signaling. This was discovered through the use of the chemicals DCMU and DBMIB, which block electron transport before and after the PQ pool in the PET chain respectively. This results in the accumulation of mostly oxidised PQ pool in DCMU-treated plants and mostly reduced PQ in DBMIB-treated ones, mimicking the effect of low- and high-light intensities respectively. Under a combination of DCMU and light treatments, almost 300 nuclear-encoded genes were found to be specifically regulated by the redox state of the PQ pool.27
However, chemical treatment becomes a far more powerful tool when coupled with mutagenic screens. The involvement of the GUN loci in modulating retrograde signaling was first elucidated through the differential response of gun1–gun5 to chemical treatment. These mutants express nuclear photosynthetic genes such as Light Harvesting Complex B (LHCB) genes despite norflurazon treatment,63 suggesting an impairment or alteration in retrograde signaling in these mutants
Excess light (EL) stress
In the search for retrograde signals, another method routinely used to perturb chloroplast function to induce operational control is the application of EL stress onto plants. EL stress is caused by absorption of light energy in excess of the plant's photosynthetic capacity;64 in nature this occurs on a daily basis as plants are exposed to fluctuating light intensities from the sun. Similar to other abiotic stresses, EL causes a decrease in photosynthetic capacity and a concomitant increase in ROS production. Accordingly, this photo-oxidative stress from EL also induces the generation of retrograde signals and the corresponding changes in nuclear gene expression. For example, the ROS scavenger APX2 shows strong induction within 15 minutes of EL stress.65 The cpSRP43 protein, which facilitates assembly of light-harvesting antennae in chloroplasts, is also responsive to EL and inhibition of photosynthesis.66 These findings suggest a strong relationship between retrograde signal-mediated regulation and light-responsive genes.
The use of EL in mutant screens has also highlighted the interaction of photoreceptors with retrograde signaling pathways.26 Photoreceptors, such as phytochromes and cryptochromes, are wavelength-specific cytosolic proteins. These proteins can undergo conformational changes in response to alterations in light to interact with downstream signaling partners.44 Among these photoreceptors, the protein CRY1 has been identified to regulate the activities of transcription factors that control expression of retrograde-controlled genes such as Early Light-Induced Protein 2 (ELIP2).26 Four cry mutant alleles were identified that all exhibited subtle gun phenotypes, indicating an integration between photoreceptors and photosynthesis signaling.26
However, retrograde signaling can also proceed via photoreceptor-independent pathways,67 and thus it is likely that signals from the chloroplast regulate only a subset of all light-responsive genes.27 This is illustrated in the cry1 mutant where the induction of ELIP2 by EL stress is strongly attenuated, but not that of APX2. This suggests a critical role of CRY1 in modulating ELIP2 expression but not that of the classic EL-responsive gene APX2.67
Drought Stress as a Model System for Studying Retrograde Signaling
Little is known about chloroplast to nucleus retrograde signaling in drought-stressed plants. However, 69% of genes induced EL are also induced by drought,68 suggesting a strong interconnection between the responses to these two types of stresses that occur simultaneously in most cases. Many abiotic stress responses, including that for drought, are likely to share common underlying signaling mechanisms or components.43 This is perhaps intuitively obvious considering that different abiotic stresses ranging from drought and excess light to nutrient deficiency all decrease the maximum photosynthetic capacity of the chloroplast32 which enhances ROS production,41,45 thus necessitating retrograde signaling.
However, drought stress—in particular soil-based dehydration experiments—presents a critical difference to chemical and EL systems for the study of retrograde signaling. While chloroplastic processes are indeed all perturbed in drought, chemical treatment and EL stress, this disruption typically occurs on a much faster timescale in the latter two systems. In contrast to the faster chemical treatment and EL experiments which have timescales of hours, soil-based dehydration experiments typically occur over a period of days.69,70 In mutagenic screens, measurement of the accumulation of a number of potential retrograde signals over a longer timeframe may allow better discrimination between molecules that only accumulate transiently and those which accumulation corresponds with a change in gene expression.
The activation of retrograde signaling via progressive reduction of soil water content during drought is arguably more physiologically relevant than some chemical treatments. Chemical treatments can be at doses that are lethal to the plant. Under such artificial conditions, the significance of alterations observed in retrograde signaling mutants cannot be overestimated. For instance, there are significant differences between the gene expression profiles of gun1 and wild type when treated with lincomycin. However, in untreated 7-day-old seedlings, only one transcript varies between the mutant and wild type regardless of whether or not light was present.71 In the case of EL stress, prolonged constant exposure to high light intensities may not mimic the situation in nature of some plants, where they may be exposed to high sunlight in fits and bursts. In contrast, soil-based drought is not an uncommon phenomenon to many plant species. A slow, cumulative perturbation of the chloroplast during drought is both evolutionarily and physiologically relevant. The question then is how does the chloroplast, a well-placed sensor of abiotic stress, communicate with the nucleus during drought?
During drought, there is a gradual reduction in soil water content. The adverse effect of this on plants is perhaps best illustrated by considering the changes in water potential (Ψw) of the soil relative to the plant water potential. The direction of the movement of water is driven by the gradient in Ψw between the plant cell and the soil. Due to the presence of solutes in the cytoplasm, plant cells have a more negative Ψw compared to well-watered soil (Ψw of pure water is zero). As water moves from regions of high Ψw to regions of lower Ψw, there is a net flow of water from the soil into plant cells, until equilibrium is established. During drought, the reduction in soil water content decreases soil Ψw72 to values closer to or lower than those of the Ψw of plant cells. The reduction of the Ψw gradient during drought affects water uptake by the plant, leading to dehydration and eventual death should an effective response against drought be absent.73
A reduction in soil water availability is first sensed by the root system. The phytohormone abscisic acid (ABA), one of the major root-to-shoot stress signals, is then conveyed from the root to other parts of the plant74 to initiate a response. Plant responses to drought can be broadly categorized as either drought avoidance or tolerance.73 Drought avoidance is characterized by maintaining tissue Ψw and water content close to unstressed levels by increasing water up-take and limiting water loss, at the expense of growth of photosynthetic tissue. On the other hand, tolerance to drought involves acclimation responses that allow the plant to maintain high water content and photosynthesis at lower Ψw. Nevertheless, a common feature of both types of drought response is that they invoke a suite of physiological modifications. In drought avoidance a high Ψw is maintained via stomatal closure followed by an increase in root growth, and hence the root/shoot ratio. In drought tolerance, physiological changes including intracellular accumulation of solutes to raise the osmotic gradient, alteration of cell wall permeability to water and accumulation of membrane-stabilizing proteins such as dehydrins.75 These changes in metabolism and physiology allow plants to survive with less water.
These physiological changes occur as a result of large-scale alterations in gene expression in response to drought. Microarray analysis of 8,100 genes in Arabidopsis plants under drought stress by Kreps et al.76 revealed changes in gene expression by two-fold or greater for 1,008 genes, of which 46% were upregulated. Comparison between different microarray experiments for drought stress identified a group of co-regulated genes. These include genes involved in cellular metabolism, cellular transport, signal transduction and transcriptional regulation; as well as those coding for gene products of unknown function.77 In contrast, genes coding for enzymes involved in cell wall biosynthesis and expansion were downregulated,77 which is consistent with the reduction of growth during drought.73
It is very likely that retrograde signaling is involved in the plant drought response. The first instance is when the plant initiates drought avoidance in shifting to a water-saving strategy by triggering stomatal closure. Ironically, while this enables the plant to reduce water loss via transpiration through stomata, it also impairs the uptake of carbon dioxide (CO2).40 Reduction in CO2 uptake leads to limited CO2 fixation in photosynthesis and thus impacts on NADP+ regeneration through the Calvin cycle. As discussed earlier, this triggers enhanced production of ROS.45 The involvement of ROS in signaling pathways during drought is well documented (reviewed in ref. 40); however, what is unclear is if ROS accumulation results in the accumulation of other chloroplast-localized molecules which may then act as retrograde signals.
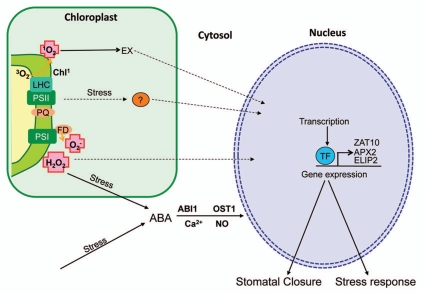
The stomatal closure-induced reduction in NADP+ regeneration through the Calvin cycle will naturally impact on photosynthesis. As drought severity increases, prolonged stomatal closure will cause a dramatic reduction in photosynthetic efficiency. This can be measured by quantifying the availability of the reaction centers in Photosystem II, for example using chlorophyll fluorescence.78 The decrease in photosynthetic efficiency may also impact on other chloroplastic processes. For instance, pathways which utilize the NADP+/NADPH pools, such as FA biogenesis,79 will be perturbed. The disruption of multiple processes in the chloroplast during drought would therefore necessitate one or more retrograde signals for the plant to initiate compensatory mechanisms. A possible model for retrograde signaling in drought and EL stress is summarized in Figure 1.
Figure 1.
Model for retrograde signaling during drought and EL stress. Drought shares a common feature with EL stress in that both stresses induce ABA-mediated signaling, which activates transcription of stress response genes such as ZAT10, APX2 and ELIP2. The production of ROS species during drought can also induce changes in gene expression leading to a drought response. At PSII, 1O2 accumulation can induce programmed cell death; this is also regulated by EXECUTER (EX), but the pathway beyond the chloroplast is unknown. ROS produced at PSI also influence gene transcription. O2− accumulation can influence gene transcription but the mechanism is unclear. While the role of H2O2 in stress response, particularly in ABA-signaling, is well-studied, it is unknown if H2O2 can directly move to the nucleus to influence transcription. Another interesting possibility is that retrograde signals may actually be metabolites normally regarded as by-products and broken down by catabolic enzymes; these molecules may accumulate during stress and influence nuclear transcription (question mark). All dashed lines indicate areas that require more research.
Towards Identifying Retrograde Signals in the Plant Drought Response
A potential strategy for identifying retrograde signals is to employ a forward genetic screen to identify mutants defective or enhanced in retrograde signaling. Consider a hypothetical mutant defective in retrograde signaling. Ideally this mutant should have perturbations in either chloroplast function or morphology, since retrograde signaling has been strongly linked to photosynthesis4 and chloroplast biogenesis.17,26 In addition, the mutant should display alterations in gene expression, consistent with the role of retrograde signaling in modulating gene expression.4,17 Perturbations in the chloroplast and gene expression may also combine to cause changes in leaf morphology and overall development.
Given that retrograde signaling also likely occurs in response to perturbation of chloroplast function during drought, the same mutant may also exhibit a significantly altered drought response, which may translate to either enhanced sensitivity or tolerance to drought. This change in drought sensitivity could be due to modifications in various drought response properties including altered stomatal conductance,80 accumulation or loss of osmoprotectants,81 changes in expression of drought-response genes69 and altered uptake of water from roots.82–84 Since abiotic stress responses in the plant are likely to be integrated43 and 69% of the drought-responsive genes are also activated in EL stress,68 this mutant may also show differential sensitivity to EL and it should exhibit a change in EL-induced gene expression. Indeed, an EL screen for mutations perturbed in retrograde signaling identified 13 lesions, of which 4 showed altered degrees of drought tolerance.43,85 Indeed, one of these had already been identified in abiotic stress screens.69,86 Thus, the converse screen or a systematic analysis of mutations that alter drought stress signaling may yield new insights into retrograde signaling.
Pfannschmidt in a recent review87 raised an interesting point in that the ‘retrograde signals’ may simply be metabolites accumulating to a certain threshold recognized by the cell as ‘metabolite signatures’, which then triggers changes in gene expression. At the same time, the role of by-products in negative feedback regulation of various chloroplast-localized pathways is well-documented. Building on from these observations, another interesting possibility would be that by-products of chloroplastic processes, with hitherto no assigned functions, might accumulate during stress and participate in retrograde signaling. Therefore, it is possible that retrograde signaling is altered in mutants lacking certain catabolic enzymes that degrade by-products of the numerous chloroplastic processes. In such mutants, accumulation of their substrates could result in amplified retrograde signaling and possibly the phenotypes discussed above. Nevertheless, if one were to go by the strict definition of retrograde signals, such substrates would have to be able to interact directly with key components of gene expression such as transcription factors, protein kinases and RNA processing enzymes.
High-throughput metabolite profiling of retrograde signaling mutants may assist the search for retrograde signals. While metabolomics is still a relatively new field compared to transcriptomics, high-throughput methods are emerging. Many different techniques, mostly variants of mass spectrometry that can study the metabolome of different organisms, including plants,88,89 yeast,90 mice and humans91 with high sensitivity and efficiency have been described in the past few years. As sensitivity of metabolomic techniques increase, so does the probability of finding retrograde signals that may be present at low concentrations.
Nevertheless, careful interpretation of the biological significance of results from metabolite profiling is required. This is aptly illustrated in studies by Brautigam et al.92 where a PSI-II light shift experiment, which alters the redox state of the chloroplast, resulted in a transient increase in intracellular sucrose concentration. At the first glance this could have been easily interpreted as a retrograde signal participating in operational control. However, detailed investigation by the authors combining transcript studies of photosynthetic genes, measurement of starch accumulation and mutant analysis led to the conclusion that this transient sucrose accumulation has a different biological role and is unlikely to be a retrograde signal.92
A concurrent proteomic approach may also complement existing and new strategies, including drought. While the literature contains relatively few examples of the direct involvement of post-translational modification in retrograde signaling, some findings are instructive. For instance, analysis of the maize chloroplast proteome identified seven chloroplast-localized proteins, including an unknown protein, that were post-translationally modified during chloroplast biogenesis.93 These proteins, and the metabolic network in which they are located, would therefore be prime targets for further investigation into signals in biogenic control. Of course, whether such post-translational modifications actually represent the starting point or the consequence of retrograde signaling is open to debate; nevertheless they represent an additional focal point in the search for retrograde signals. For example, would analysis of the chloroplast proteome during drought reveal post-translationally modified proteins that help to generate retrograde signals or act as retrograde signals themselves?
Concluding Remarks
Retrograde signaling is undeniably an important facet of plant biology, coordinating the multitude of processes between different cellular compartments. Chloroplast-to-nucleus communication, in particular, is vital considering the numerous important processes occurring in the chloroplast. Despite major advances in our understanding of retrograde signaling over the past few decades, however, much uncertainty remains. What are the actual retrograde signals? How do perturbations in chloroplastic processes translate into accumulation of retrograde signals and changes in nuclear gene expression? If indeed retrograde signals need to accumulate to act, does this accumulation work by a threshold, “on/off” mechanism? Last but not least, could by-products of chloroplastic processes act as retrograde signals? These questions and more will need to be addressed in order for us to elucidate the workings of retrograde signaling. Drought stress combined with mutagenesis may present a viable alternative to the ‘traditional’ chemical and EL experiments in helping us answer these questions.
Acknowledgements
This work was funded by the Australian Research Council Centre of Excellence in Plant Energy Biology (CE0561495).
Abbreviations
- FA
fatty acid
- ABA
abscisic acid
- Mg-protoPIX
Mg-protoporphyrin IX
- GUN
GENOMES UNCOUPLED
- ROS
reactive oxygen species
- H2O2
hydrogen peroxide
- 1O2
singlet oxygen
- O2·−
superoxide anion
- APX2
ascorbate peroxidase 2
- EL
excess light
- flu
fluorescent
- ex1
executer 1
- CuZn-SOD
copper-zinc superoxide dismutase
- PET
photosynthetic electron transport
- PQ
plastoquinone
- ELIP2
early light-induced protein 2
- Ψw
water potential
- CO2
carbon dioxide
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13758
References
- 1.Singh BK, Matthews BF. Molecular regulation of amino-acid biosynthesis in plants. Amino Acids. 1994;7:165–174. doi: 10.1007/BF00814158. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annual Review of Plant Physiol Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- 3.Abdallah F, Salamini F, Leister D. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 2000;5:141–142. doi: 10.1016/s1360-1385(00)01574-0. [DOI] [PubMed] [Google Scholar]
- 4.Pogson BJ, Woo NS, Forster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13:602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Taylor WC. Regulatory interactions between nuclear and plastid genomes. Ann Rev Plant Physiol Plant Mol Biol. 1989;40:211–233. [Google Scholar]
- 6.Surpin M, Chory J. The co-ordination of nuclear and organellar genome expression in eukaryotic cells. Essays Biochem. 1997;32:113–125. [PubMed] [Google Scholar]
- 7.Rodermel S. Pathways of plastid-to-nucleus signaling. Trends Plant Sci. 2001;6:471–478. doi: 10.1016/s1360-1385(01)02085-4. [DOI] [PubMed] [Google Scholar]
- 8.Mullineaux P, Karpinski S. Signal transduction in response to excess light: getting out of the chloroplast. Curr Opin Plant Biol. 2002;5:43–48. doi: 10.1016/s1369-5266(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 9.Strand A. Plastid-to-nucleus signalling. Curr Opin Plant Biol. 2004;7:621–625. doi: 10.1016/j.pbi.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Beck CF. Signaling pathways from the chloroplast to the nucleus. Planta. 2005;222:743–756. doi: 10.1007/s00425-005-0021-2. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis P. Intracellular signalling: Chloroplast backchat. Curr Biol. 2007;17:552–555. doi: 10.1016/j.cub.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Larkin RM, Ruckle ME. Integration of light and plastid signals. Curr Opin Plant Biol. 2008;11:593–599. doi: 10.1016/j.pbi.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Dyall SD, Brown MT, Johnson PJ. Ancient invasions: From endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Juez E, Pyke KA. Plastids unleashed: their development and their integration in plant development. Int J Dev Biol. 2005;49:557–577. doi: 10.1387/ijdb.051997el. [DOI] [PubMed] [Google Scholar]
- 15.Kopriva S. Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot. 2006;97:479–495. doi: 10.1093/aob/mcl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, et al. Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog Lipid Res. 2010;49:128–158. doi: 10.1016/j.plipres.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Waters MT, Langdale JA. The making of a chloroplast. EMBO J. 2009;28:2861–2873. doi: 10.1038/emboj.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dun EA, Brewer PB, Beveridge CA. Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci. 2009;14:364–372. doi: 10.1016/j.tplants.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Milborrow BV. The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J Exper Bot. 2001;52:1145–1164. [PubMed] [Google Scholar]
- 20.Chinnusamy V, Gong ZZ, Zhu JK. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol. 2008;50:1187–1195. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benech-Arnold RL, Enciso S, Sanchez RA, Carrari F, Perez-Flores L, Iusem N, et al. Involvement of ABA and GAs in the regulation of dormancy in developing sorghum seeds. In: Black M, Bradford KJ, Vazquez-Ramos J, editors. Seed biology: advances and applications. Proceedings of the Sixth International Workshop on Seeds. Cambridge: Cabi Publishing; 2000. pp. 101–111. [Google Scholar]
- 22.Benning C. A role for lipid trafficking in chloroplast biogenesis. Prog Lipid Res. 2008;47:381–389. doi: 10.1016/j.plipres.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Kubis S, Patel R, Combe J, Bedard J, Kovacheva S, Lilley K, et al. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell. 2004;16:2059–2077. doi: 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao H, Sage TL, Osteryoung KW. FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc Natl Acad Sci. 2006;103:6759–6764. doi: 10.1073/pnas.0507287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pontier D, Albrieux C, Joyard J, Lagrange T, Block MA. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. J Biol Chem. 2007;282:2297–2304. doi: 10.1074/jbc.M610286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruckle ME, DeMarco SM, Larkin RM. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell. 2007;19:3944–3960. doi: 10.1105/tpc.107.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fey V, Wagner R, Brautigam K, Wirtz M, Hell R, Dietzmann A, et al. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem. 2005;280:5318–5328. doi: 10.1074/jbc.M406358200. [DOI] [PubMed] [Google Scholar]
- 28.Oelmuller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H. Expression of nuclear genes as affected by treatments acting on the plastids. Planta. 1986;168:482–492. doi: 10.1007/BF00392267. [DOI] [PubMed] [Google Scholar]
- 29.Mugford SG, Yoshimoto N, Reichelt M, Wirtz M, Hill L, Mugford ST, et al. Disruption of adenosine-5′-Phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell. 2009;21:910–927. doi: 10.1105/tpc.109.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohlrogge JB, Jaworski JG. Regulation of fatty acid synthesis. Ann Rev Plant Physiol Plant Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez AP, Strand A. Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr Opin Plant Biol. 2008;11:509–513. doi: 10.1016/j.pbi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Li ZR, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 33.Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 34.Ankele E, Kindgren P, Pesquet E, Strand A. In vivo visualization of Mg-ProtoporphyrinIX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell. 2007;19:1964–1979. doi: 10.1105/tpc.106.048744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- 36.Zhang DP. PLANT SCIENCE: Signaling to the Nucleus with a Loaded GUN. Science. 2007;316:700–701. doi: 10.1126/science.1142703. [DOI] [PubMed] [Google Scholar]
- 37.Susek RE, Ausubel FM, Chory J. Signal-transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene-expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 38.Moulin M, McCormac AC, Terry MJ, Smith AG. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci. 2008;105:15178–15183. doi: 10.1073/pnas.0803054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz De Carvalho MH. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Sig and Behav. 2008;3:156–165. doi: 10.4161/psb.3.3.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker NR. A possible role for photosystem II in environmental perturbations of photosynthesis. Physiol Plant. 1991;81:563–570. [Google Scholar]
- 42.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 43.Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, et al. A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 2006;29:269–281. doi: 10.1111/j.1365-3040.2005.01419.x. [DOI] [PubMed] [Google Scholar]
- 44.Foyer CH, Noctor G. Redox Regulation in photosynthetic organisms: Signaling, acclimation and practical implications. Antioxid Redox Signal. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- 45.Smirnoff N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytologist. 1993;125:27–58. doi: 10.1111/j.1469-8137.1993.tb03863.x. [DOI] [PubMed] [Google Scholar]
- 46.Desikan R, Clarke A, Hancock JT, Neill SJ. H2O2 activates a MAP kinase-like enzyme in Arabidopsis thaliana suspension cultures. J Experiment Bot. 1999;50:1863–1866. [Google Scholar]
- 47.Grant JJ, Yun BW, Loake GJ. Oxidative burst and cognate redox signalling reported by luciferase imaging: identification of a signal network that functions independently of ethylene, SA and Me-JA but is dependent on MAPKK activity. Plant J. 2000;24:569–582. doi: 10.1046/j.1365-313x.2000.00902.x. [DOI] [PubMed] [Google Scholar]
- 48.Neill S. Interactions between abscisic acid, hydrogen peroxide and nitric oxide mediate survival responses during water stress. New Phytologist. 2007;175:4–6. doi: 10.1111/j.1469-8137.2007.02112.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang PT, Song CP. Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytologist. 2008;178:703–718. doi: 10.1111/j.1469-8137.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- 50.Yun KY, Park MR, Mohanty B, Herath V, Xu FY, Mauleon R, et al. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. 2010:10. doi: 10.1186/1471-2229-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding MQ, Hou PC, Shen X, Wang MJ, Deng SR, Sun J, et al. Salt-induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt-sensitive poplar species. Plant Mol Biol. 2010;73:251–269. doi: 10.1007/s11103-010-9612-9. [DOI] [PubMed] [Google Scholar]
- 52.op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim CH, Danon A, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in arabidopsis. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meskauskiene R, Nater M, Goslings D, Kessler F, Op den Camp R, Apel K. FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee KP, Kim C, Landgraf F, Apel K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Nat Acad Sci USA. 2007;104:10270–10275. doi: 10.1073/pnas.0702061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner D, Przybyla D, Op Den Camp R, Kim C, Landgraf F, Keun PL, et al. The genetic basis of singlet oxygen-induced stress response of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 56.Scarpeci TE, Zanor MI, Carrillo N, Mueller-Roeber B, Valle EM. Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: A focus on rapidly induced genes. Plant Mol Biol. 2008;66:361–378. doi: 10.1007/s11103-007-9274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizhsky L, Liang H, Mittler R. The water-water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem. 2003;278:38921–38925. doi: 10.1074/jbc.M304987200. [DOI] [PubMed] [Google Scholar]
- 58.Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yabuta Y, Maruta T, Yoshimura K, Ishikawa T, Shigeoka S. Two distinct redox signaling pathways for cytosolic APX induction under photooxidative stress. Plant Cell Physiol. 2004;45:1586–1594. doi: 10.1093/pcp/pch181. [DOI] [PubMed] [Google Scholar]
- 60.Rapp JC, Mullet JE. Chloroplast transcription is required to express the nuclear genes rbcS and cab. Plastid DNA copy number is regulated independently. Plant Mol Biol. 1991;17:813–823. doi: 10.1007/BF00037063. [DOI] [PubMed] [Google Scholar]
- 61.Sullivan JA, Gray JC. Multiple plastid signals regulate the expression of the pea plastocyanin gene in pea and transgenic tobacco plants. Plant J. 2002;32:763–774. doi: 10.1046/j.1365-313x.2002.01464.x. [DOI] [PubMed] [Google Scholar]
- 62.Chamovitz D, Pecker I, Hirschberg J. The molecular-basis of resistance to the herbicide norflurazon. Plant Mol Biol. 1991;16:967–974. doi: 10.1007/BF00016069. [DOI] [PubMed] [Google Scholar]
- 63.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horton P, Ruban AV, Walters RG. Regulation of light harvesting in green plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:655–684. doi: 10.1146/annurev.arplant.47.1.655. [DOI] [PubMed] [Google Scholar]
- 65.Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, et al. Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell. 2007;19:4091–4110. doi: 10.1105/tpc.106.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klenell M, Morita S, Tiemblo-Olmo M, Muhlenbock P, Karpinski S, Karpinska B. Involvement of the chloroplast signal recognition particle cpSRP43 in acclimation to conditions promoting photooxidative stress in Arabidopsis. Plant Cell Physiol. 2005;46:118–129. doi: 10.1093/pcp/pci010. [DOI] [PubMed] [Google Scholar]
- 67.Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of arabidopsis to high irradiance. Plant Physiol. 2007;144:1391–1406. doi: 10.1104/pp.107.098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimura M, Yamamoto YY, Seki M, Sakurai T, Sato M, Abe T, et al. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem Photobiol. 2003;77:226–233. doi: 10.1562/0031-8655(2003)077<0226:ioagrb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 69.Wilson PB, Estavillo GM, Field KJ, Pornsiriwong W, Carroll AJ, Howell KA, et al. The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 2009;58:299–317. doi: 10.1111/j.1365-313X.2008.03780.x. [DOI] [PubMed] [Google Scholar]
- 70.Alcazar R, Planas J, Saxena T, Zarza X, Bortolotti C, Cuevas J, et al. Putrescine accumulation confers drought tolerance in transgenic Arabidopsis plants overexpressing the homologous Arginine decarboxylase 2 gene. Plant Physiol Biochem. 2010;48:547–552. doi: 10.1016/j.plaphy.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 71.Cottage AJ, Mott EK, Wang JH, Sullivan JA, MacLean D, Tran L, et al. GUN1 (GENOMES UNCOUPLED1) Encodes a pentatricopeptide repeat (PPR) protein involved in plastid protein synthesis-responsive retrograde signaling to the nucleus. In: Allen JF, editor. Photosynthesis Energy from the Sun. Springer; 2008. pp. 1202–1205. [Google Scholar]
- 72.Kramer PJ, Bwwwoyle JS. Water Relations of Plants and Soils. San Diego: Academic Press; 1995. [Google Scholar]
- 73.Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu JH, Zhu JK. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45:523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 74.Jiang F, Hartung W. Long-distance signalling of abscisic acid (ABA): the factors regulating the intensity of the ABA signal. J Exper Bot. 2007:127. doi: 10.1093/jxb/erm127. [DOI] [PubMed] [Google Scholar]
- 75.Bravo LA, Gallardo J, Navarrete A, Olave N, Martinez J, Alberdi M, et al. Cryoprotective activity of a cold-induced dehydrin purified from barley. Physiol Plant. 2003;118:262–269. [Google Scholar]
- 76.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J Exper Bot. 2004;55:2331–2341. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- 78.Woo NS, Badger MR, Pogson BJ. A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods. 2008;4:27. doi: 10.1186/1746-4811-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harwood JL. Fatty acid metabolism. Ann Rev Plant Physiol Plant Mol Biol. 1988;39:101–138. [Google Scholar]
- 80.Yoo CY, Pence HE, Hasegawa PM, Mickelbart MV. Regulation of transpiration to improve crop water use. CRC Crit Rev Plant Sci. 2009;28:410–431. [Google Scholar]
- 81.Kalamaki MS, Merkouropoulos G, Kanellis AK. Can ornithine accumulation modulate abiotic stress tolerance in Arabidopsis? Plant Signal Behav. 2009;4:1099–1101. doi: 10.4161/psb.4.11.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu HS, Li FM, Xu H. Deficiency of water can enhance root respiration rate of drought-sensitive but not drought-tolerant spring wheat. Agricultural Water Management. 2004;64:41–48. [Google Scholar]
- 83.Farooq M, Wahid A, Lee DJ, Ito O, Siddique KHM. Advances in drought resistance of rice. CRC Crit Rev Plant Sci. 2009;28:199–217. [Google Scholar]
- 84.Manschadi AM, Hammer GL, Christopher JT, deVoil P. Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.) Plant Soil. 2008;303:115–129. [Google Scholar]
- 85.Rossel JB, Cuttriss A, Pogson BJ. Identifying photoprotection mutants in Arabidopsis thaliana. Methods Mol Biol. 2004;274:287–299. doi: 10.1385/1-59259-799-8:287. [DOI] [PubMed] [Google Scholar]
- 86.Xiong LM, Lee BH, Ishitani M, Lee H, Zhang CQ, Zhu JK. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001;15:1971–1984. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pfannschmidt T. Plastidial retrograde signalling—a true plastid factor or just metabolite signatures? Trends Plant Sci. 2010;15(8):427–423. doi: 10.1016/j.tplants.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 88.Sawada Y, Akiyama K, Sakata A, Kuwahara A, Otsuki H, Sakurai T, et al. Widely targeted metabolomics based on large-Scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 2009;50:37–47. doi: 10.1093/pcp/pcn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: An Application for Hormone Profiling in Oryza sativa. Plant Cell Physiol. 2009;50:1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ewald JC, Heux Sp, Zamboni N. High-throughput quantitative metabolomics: Workflow for cultivation, quenching and analysis of yeast in a multiwell format. Anal Chem. 2009;81:3623–3629. doi: 10.1021/ac900002u. [DOI] [PubMed] [Google Scholar]
- 91.Han J, Danell R, Patel J, Gumerov D, Scarlett C, Speir J, et al. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics. 2008;4:128–140. doi: 10.1007/s11306-008-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brautigam K, Dietzel L, Kleine T, Stroher E, Wormuth D, Dietz KJ, et al. Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell. 2009;21:2715–2732. doi: 10.1105/tpc.108.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lonosky PM, Zhang X, Honavar VG, Dobbs DL, Fu A, Rodermel SR. A proteomic analysis of maize chloroplast biogenesis. Plant Physiol. 2004;134:560–574. doi: 10.1104/pp.103.032003. [DOI] [PMC free article] [PubMed] [Google Scholar]