Abstract
The interaction between a bacterial pathogen and its potential plant host develops from a complex combination of bacterial and plant elements, which determines either the establishment of resistance or the development of disease. The use of virulence assays based on competitive index in mixed infections constitutes a powerful tool for the analysis of bacterial virulence factors. In this work, we describe how the use of competitive index assays also constitutes an alternative approach for the analysis of plant immunity, to determine the contribution of different elements to bacterial recognition or immunity signaling.
Key words: competitive index, mixed infections, pathogen, plant immunity, defence response, effector-triggered immunity
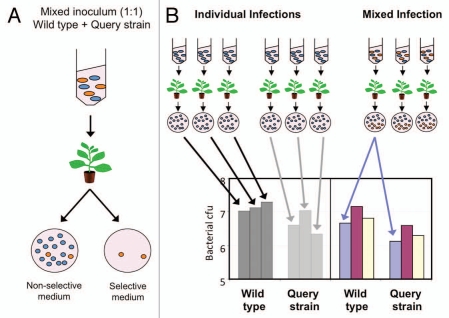
The type III secretion system (T3SS) allows Gram negative bacterial pathogens to deliver a set of effector proteins into the host cell. The plant pathogen Pseudomonas syringae employs a large inventory of type III-secreted effectors (T3SEs) to suppress plant immunity. Individual mutation of effector genes has traditionally failed to provide a relevant virulence phenotype, a fact generally associated to a high degree of functional redundancy between T3SEs, which also hinders the characterisation of effector activities within the plant cell. This problem has led researchers to use alternative approaches to overcome functional redundancy, including the generation of polymutants lacking several effector genes, ectopic expression of effectors in heterologous strains lacking the corresponding homolog, as well as the generation of transgenic plants expressing a given effector.1 We have previously established that the use of competitive index in mixed infections provides an accurate and sensitive manner of establishing virulence phenotypes for single effector mutants for which other assays have failed,2 thus providing an alternative to previously used approaches for the analysis of effector function within the context of the infection. This increase in sensitivity and accuracy is due to the direct comparison between growth of the co-inoculated strains within the same infection (Fig. 1), which replicate as they would in individual infections under the appropriate experimental settings.2
Figure 1.
Basis for the increased sensitivity and acuracy of CI assays. A mixed inoculum with equal amounts of wild type and query strain is inoculated within the same plant (A), allowing a direct comparison between the replication values of both strains within the same infection (B). On the contrary, in regular individual infections, the values obtained from different plants have to be pulled first and compared afterwards (B), thus accumulating experimental and plant-to-plant variation.
Plant immunity can be triggered by a group of conserved microbial molecules known as PAMPs (pathogen-associated molecular patterns). PAMP-triggered immunity (PTI) can be suppressed by effectors which in turn can be recognized by nucleotide binding-leucine rich repeat (NB-LRR) proteins, encoded by resistance genes or R genes.3,4 Detection of such effectors by NB-LRR proteins determines effector-triggered immunity (ETI),3 an amplified version of PTI, which usually crosses the threshold inducing the hypersensitive response (HR), a localized cell death response. Furthermore, bacteria have evolved effectors that suppress ETI, some of which can in turn be recognized by the plant, thus triggering a secondary ETI.4 Selection favors new plant NB-LRRs that can recognize such secondary, newly acquired effectors.
R-gene-mediated defences are usually associated with the accumulation of salycilic acid (SA),5 although SA-independent pathways such as that dependent on EDS1 (enhanced disease susceptibility-1),6 can also mediate ETI and trigger an HR.7 Although regulating independent pathways, EDS1 and SA have been recently described to function redundantly to regulate R-gene-mediated signaling.8
The Competitive Index Angle
Our work applying CI assays has been based on the knowledge generated from the different applications of this method in animal pathogens, and has proven that it is possible to adapt this methodology to the analysis of bacterial growth in several plant hosts of, at least, two bacterial species of major relevance among microbe-plant interactions: Pseudomonas syringae and Ralstonia solanacearum.2,9 The versatility that mixed infections have demonstrated as a methodology for genetic analysis of virulence suggests that they are potentially adaptable to other models of bacteria-plant interactions. So far, we have taken advantage of the high sensitivity and accuracy offered by CI assays to determine the contribution of many effector proteins to bacterial growth within the host plant.2,9,10 CI and CI-based assays have also been used to carry out genetic analysis by comparing the virulence of co-inoculated single and/or multiple mutants.11,12 We have gone beyond its use in animal pathogens in finding applications for CI analysis, using it to identify novel translocated effectors, by using a modification of a reporter-based translocation assay.13 In a recent report, we have also used competitive index assays to analyse functional relationships between T3SEs within the plant cell, showing that HopZ1a triggers a partially additive plant defence response to that triggered by the well characterized effectors AvrRpt2, AvrRpm1 and AvrRps4.14 In addition, we applied CI assays to rule out the hypothesis of these partially additive defences being due to the signaling pathways sharing common elements, demonstrating that HopZ1-triggered immunity is indepedent of SA, EDS1, jasmonic acid (JA) and ethyene (ET)-dependent pathways. These results, in addition to other phenotypic and molecular assays, allowed us to establish that HopZ1a suppresses ETI. In this report, we were also able to detect a reduction in the defence response triggered by AvrRpt2 in eds1 mutant plants,14 revealing the quantitative contribution of EDS1 to RPS2-mediated signaling. It is noteworthy that this contribution had only been identified thus far through the use of double mutant plants impaired in SA and EDS1-mediated pathways.8
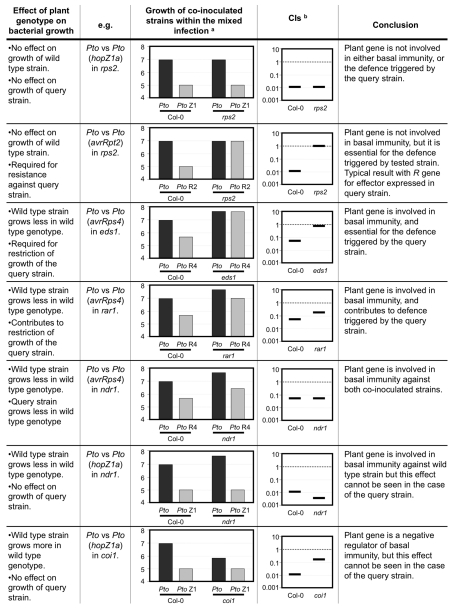
The use of CIs to analyse the contribution of particular plant genes in determining the plant-pathogen interaction has proven to be very straightforward when the plant genotype assayed does not affect the growth of the wild type strain used as a reference within the CI. In such case, the differences between the CI values obtained for the same query-wild type strain combination, when assayed in different plant genotypes, do reflect the contribution of those plant genotypes to the growth of the query strain. However, when growth of the wild type strain is affected by the plant genotype being tested, a more complex analysis of the CI results is required. Plant genotypes displaying enhanced susceptibility to the wild type strain can give smaller CI values without actually facilitating the growth of the query strain tested (Fig. 2). On the other hand, plant genotypes particularly resistant to the wild type strain can give CI values closer to one, without being actually compromised in their resistance to the tested strain (Fig. 2). Thus, when applying CI assays to analyse the role of a plant genotype in the differential growth of the query bacterial strain, it is necessary to consider carefully the actual bacterial cfu numbers obtained for each strain within the mixed infection in each of the plant genotypes assayed, to make sure that the plant genotype is actually affecting the growth of the tested strain, and that the CI output is not misleading as a consequence of an altered growth behavior in a particular plant genotype of the wild type strain used as an internal control (Fig. 2). The possible outputs for CI assays of plant genotypes, illustrated with examples that we have encountered throughout our work, are represented in Figure 2. This figure includes CI and cfu values for each type of interaction, together with their particular interpretations, thus providing the guidelines for the use of CI assays to analyse the role of plant genes in plant-pathogen interactions. As we show here, the use of CI assays in different plant genotypes, more complex to interpret than standard CI assays but equally straightforward to perform, can constitute a powerful tool, complementary to molecular analysis, to elucidate the contribution of different plant signaling elements in the context of a defence response.
Figure 2.
Mixed Infection-based analysis of the contribution of a given plant genotype to the defence triggered by a query bacterial strain. aBacterial numbers are given in log (cfu/cm2). bBars represent competitive index values representative of the tested strains in the corresponding plant genotypes. Both bacterial numbers and competitive index values are indicative of typical values for easier comparison. Accurate numbers are presented in reference 14.
Acknowledgements
We are grateful to J. Ruiz-Albert for critical reading of the manuscript. The work was supported by Project Grants from the Ministerio de Educación y Ciencia (BIO2006-00673), and Ministerio de Ciencia e Innovación (BIO2009-11516) (Spain) to C.R. Beuzón. The work was co-funded by Fondos Europeos de Desarrollo Regional (FEDER).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13843
References
- 1.Cunnac S, Lindeberg M, Collmer A. Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr Opin Microbiol. 2009;12:53–60. doi: 10.1016/j.mib.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Macho AP, Zumaquero A, Ortiz-Martín I, Beuzón CR. Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae-plant interactions. Mol Plant Pathol. 2007;8:437–450. doi: 10.1111/j.1364-3703.2007.00404.x. [DOI] [PubMed] [Google Scholar]
- 3.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 6.Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venugopal SC, Jeong RD, Mandal MK, Zhu S, Chandra-Shekara AC, Xia Y, et al. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 2009;5:1000545. doi: 10.1371/journal.pgen.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macho AP, Guidot A, Barberis P, Beuzon CR, Genin S. A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol Plant Microbe Interact. 2010;23:1197–1205. doi: 10.1094/MPMI-23-9-1197. [DOI] [PubMed] [Google Scholar]
- 10.Zumaquero A, Macho AP, Rufian JS, Beuzon CR. Analysis of the role of the type III effector inventory of Pseudomonas syringae pv. phaseolicola 1448a in interaction with the plant. J Bacteriol. 2010;192:4474–4488. doi: 10.1128/JB.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munkvold KR, Russell AB, Kvitko BH, Collmer A. Pseudomonas syringae pv. tomato DC3000 type III effector HopAA1-1 functions redundantly with chlorosis-promoting factor PSPTO4723 to produce bacterial speck lesions in host tomato. Mol Plant Microbe Interact. 2009;22:1341–1355. doi: 10.1094/MPMI-22-11-1341. [DOI] [PubMed] [Google Scholar]
- 12.Ortiz-Martin I, Thwaites R, Macho AP, Mansfield JW, Beuzon CR. Positive regulation of the Hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol Plant Microbe Interact. 2010;23:665–681. doi: 10.1094/MPMI-23-5-0665. [DOI] [PubMed] [Google Scholar]
- 13.Macho AP, Ruiz-Albert J, Tornero P, Beuzón CR. Identification of new type III effectors and analysis of the plant response by competitive index. Mol Plant Pathol. 2009;10:69–80. doi: 10.1111/j.1364-3703.2008.00511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macho AP, Guevara CM, Tornero P, Ruiz-Albert J, Beuzon CR. The Pseudomonas syringae effector protein HopZ1a suppresses effector-triggered immunity. New Phytol. 2010;187:1018–1033. doi: 10.1111/j.1469-8137.2010.03381.x. [DOI] [PubMed] [Google Scholar]