Abstract
Chloroplast photorelocation movement towards weak light and away from strong light is essential for plants to adapt to the fluctuation of ambient light conditions. In the previous study, we showed that blue light receptor phototropins mediated blue light-induced chloroplast movement in Arabidopsis by regulating short actin filaments localized at the chloroplast periphery (cp-actin filaments) rather than actin cables in the cytoplasm. However, the signaling pathway for the chloroplast photorelocation movement is still unclear. We also identified JAC1 (J-domain protein required for chloroplast accumulation response 1) as an essential component for the accumulation response and dark positioning in Arabidopsis. We recently determined the crystal structure of the J-domain of JAC1. The JAC1 J-domain has a positively charged surface, which forms a putative interface with the Hsc70 chaperone by analogy to that of bovine auxilin. Furthermore, the mutation of the highly conserved HPD motif in the JAC1 J-domain impaired the in vivo activity of JAC1. These data suggest that JAC1 cochaperone activity with HSC70 is essential for chloroplast photorelocation movement.
Key words: Arabidopsis, auxilin, blue light, clathrin, endocytosis, J-domain, organelle movement, phototropin
An Overview of the Molecular Components for Chloroplast Photorelocation Movement
Chloroplasts move to low-intensity light to capture photosynthetic light efficiently (accumulation response), whereas they escape from high-intensity light to avoid photodamage (avoidance response).1–3 Blue light is the most effective to induce chloroplast movement in various plant species. Blue light receptors, phototropins (phot), mediate chloroplast photorelocation movement in green plants.1,2 With a few exceptions, plants exclusively utilize actin filaments in the chloroplast motility system.1,2,4 Recent molecular genetic analyses using Arabidopsis thaliana identified various molecular components.1,2,4 phot1 and phot2 redundantly regulate the accumulation response,5 whereas phot2 alone is indispensable for the avoidance response.6,7 PMI1 (plastid movement impaired 1) and PMI2 are plant-specific proteins necessary for the efficient chloroplast movement.8,9 CHUP1 (chloroplast unusual positioning 1) is a chloroplast outer envelope protein capable of interacting with F-actin, G-actin and profilin in vitro.10–12 Two kinesin-like proteins, KAC1 and KAC2, are essential for chloroplast photorelocation movement and for the anchoring to the plasma membrane.13 Recently, we revealed that short actin filaments on chloroplasts (named as cp-actin filaments), but not cytoplasmic actin filaments, mediated chloroplast photorelocation movement and the anchoring to the plasma membrane.14 Phototropins regulate blue light-induced reorganization of cp-actin filaments.14 Both chup1 and kac1kac2 mutant plants lack cp-actin filaments,13,14 suggesting that CHUP1 and KACs play an important role in generation and/or maintenance of cp-actin filaments. However, specific signaling pathways regulating the accumulation and avoidance responses remain to be eluciated.
Structure and Activity of JAC1 J-domain Reveal the Possible Involvement of Hsc70 Cochaperone in Chloroplast Movement
We previously identified a jac1 (J-domain protein required for chloroplast accumulation response 1) mutant lacking chloroplast accumulation response but holding the avoidance response.15 Thus, the understanding of the JAC1 function could uncover the signaling pathway for the accumulation response. JAC1 has a J-domain at the C-terminus. J-domain is a 70-amino acid residue domain containing a highly conserved His-Pro-Asp tripetide sequence and found in the Hsp40 (heat shock protein 40 kDa) family members, including E. coli DnaJ and human Hdj1.16–18 J-domain-containing proteins (J-proteins) can be divided into three types by a sequence-based classification system.17,18 Type I J-proteins belong to the bona fide Hsp40 family and have the same domain organization as DnaJ (an N-terminal J-domain, a glycine-rich region, zinc-finger domains and a C-terminal domain). Type II J-proteins are similar to type I, but lack zinc-finger domains. In contrast, type III J-proteins contain a J-domain somewhere in the proteins, and lack other domains found in the type I and type II J-proteins. Most of the type III J-proteins have domains that are not found in type I and type II J-proteins. J-proteins exert their cellular functions via their J-domains by regulating the activity of the Hsp70 (heat shock protein 70) partners.16–18 JAC1 is one of the seven Arabidopsis proteins similar to a type III J-protein, auxilin.15
Auxilin functions as a cofactor of Hsc70 (heat shock protein cognate 70), which is a clathrin-coated vesicle uncoating factor during endocytosis.19 Auxilin J-domain mediates clathrin uncoating by recruiting and activating Hsc70.19 Moreover, auxilin and Hsc70 play multiple roles in clathrin-mediated endocytosis; chaperoning clathrin and adaptors after the dissociation from clathrin-coated vesicles, and in constricting the clathrin-coated vesicles.19 Although the sequence similarity of the JAC1 J-domain to the auxilin J-domain suggests that JAC1 could mediate chloroplast photorelocation movement through a similar molecular mechanism of auxilin, the involvement of Hsc70 and clathrin-mediated endocytosis in chloroplast movement has not been reported.
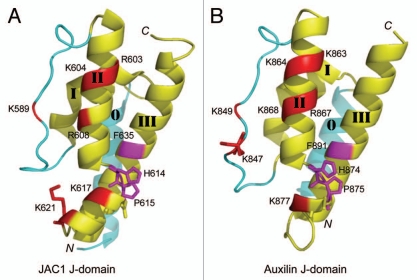
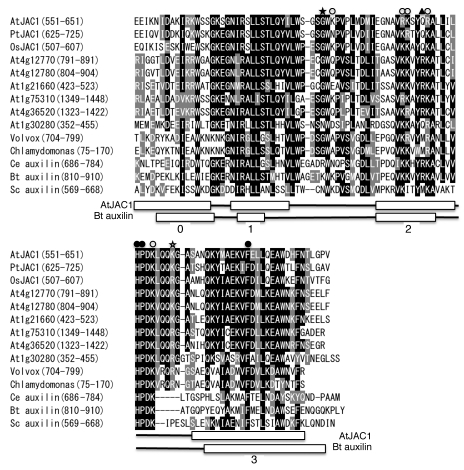
To address the structural similarity between the JAC1 and auxilin J-domains, we determined the crystal structure of the JAC1 J-domain at a 1.8 Å resolution (Fig. 1).20 The JAC1 J-domain consists of a three-helix bundle (composed of helices I, II and III) similar to the J-domains in other Hsp40 family proteins,16–18 and an additional N-terminal helix (helix 0), which uniquely exists in bovine auxilin J-domain (Fig. 1).21,22 The H874 and P875 residues of the HPD motif together with F891 in helix III of bovine auxilin J-domain mediate hydrophobic interactions with the nucleotide-binding domain of Hsc70.22 The position and side-chain orientation of the corresponding hydrophobic residues of the JAC1 J-domain are the same as those of the auxilin J-domain (Figs. 1 and 2).20 Basic residues of the loop between helices I and II (K847 and K849), the helix II (K863, K864, R867 and K868), and the HPD loop (H874 and K877 between helices II and III) of auxilin J-domain form a part of the positively charged surface, which has been proposed to be involved in the ionic interaction of the auxilin J-domain with Hsc70 (Figs. 1 and 2).21 The corresponding basic residues are conserved in the JAC1 J-domain except for residues corresponding to auxilin K847 and R867 (Fig. 2). Particularly, K847 is essential for the interaction between auxilin and Hsc70 in vitro.22 However, this essential lysine residue is absent in the J-domains of all green plant auxilin-like J-proteins, and is changed to glycine with a few exceptions (Fig. 2). Interestingly, visual inspection suggests that JAC1 K621 occupies a similar position to bovine auxilin K847 in the three-dimensional structure (Fig. 1).20 JAC1 K621 is located in the inserted segment in the loop between helices II and III. The inserted segment and the corresponding basic residue is found in all green plant auxilin-like J-proteins (Fig. 2). Thus, the charged surface formed by the inserted segment is thought to be widely conserved from green alga to flowering plants. In summary, although auxilin and green plant auxilin-like J-proteins utilize different loops to form the positively charged surface, they adopt similar ionic and hydrophobic interactions with Hsc70.
Figure 1.
Ribbon models of the J-domain of JAC1 (A) and bovine auxilin (B). The coordinates of the JAC1 and auxilin J-domains were from PDB entry 3AG7 and 2QWQ, respectively. Three-helix bundle (composed of helices I–III) similar to the J-domains in other Hsp40 family proteins is colored yellow, whereas the extra loop and auxilin-specific helix 0 are colored cyan. The positively charged and hydrophobic residues in the bovine auxilin that interact with Hsc70, and its corresponding residues in the JAC1 J-domain, are colored red and magenta, respectively. The side-chains of the HPD motif and, the essential lysine (K847) for the activity in the bovine auxilin and the alternative lysine (K621) in the JAC1 J-domain are superimposed on each ribbon models.
Figure 2.
Amino acid sequence alignment of the J-domains from plant and animal auxilin family proteins. Protein sequence alignment was carried out using the ClustalW program and visualized with BOXSHADE 3.21. At, A. thaliana; Pt, Populus trichocarpa; Os, Oryza sativa; Ce, Caenorhabditis elegans; Bt, Bos taurus; Sc, Saccharomyces cerevisiae. Arabidopsis sequences were obtained from the TAIR9 Proteins database. Chlamydomonas and Volvox sequences were obtained from respective JGI genome databases. The secondary structures of JAC1,20 and bovine auxilin21 are shown (α-helices (α): white rectangles, loop (L): black lines). Basic amino acid residues forming the positively charged surface and hydrophobic residues for the interaction with Hsc70 are indicated by open and closed circles, respectively. The residue corresponding to auxilin R867 is changed into non-basic residues in angiosperm JAC1 and some auxilin-like proteins (indicated by a closed triangle). The essential lysine residue for the Hsc70 activation in auxilin and its corresponding residues of other auxilin-like proteins are indicated by a closed star. The green-plant specific basic residues contributing to the positively charged surface are indicated by an open star.
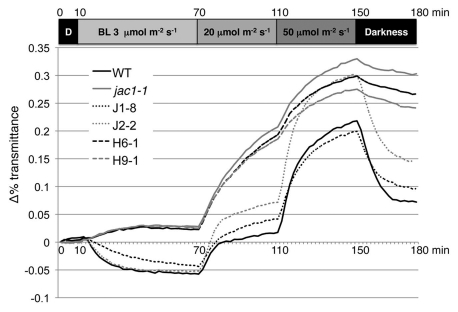
To analyze the function of the JAC1 J-domain, we constructed transgenic jac1 mutant lines expressing wild-type or HPD motif-mutated (HPD to AAA) GFP-JAC1 transgenes under the control of the JAC1 native promoter.20 The corresponding mutations in other J-proteins impaired the in vivo functions, such as the activation of their Hsp70 partners.23,24 As expected, the wild-type GFP-JAC1 fully rescued the defect of jac1 in the accumulation response, whereas HPD motif-mutated GFP-JAC1 could not at all (Fig. 3).20 Therefore, the J-domain activity is essential for the JAC1 function.
Figure 3.
Chloroplast photorelocation movement in GFP-JAC1 transgenic lines. Wild type, jac1-1, transgenic jac1 mutant lines expressing wild-type (J1-8 and J2-2) or HPD motif-mutated (H6-1 and H9-1) GFP-JAC1 transgenes were analyzed. The graph shows data from a representative experiment using six to eight leaves of each indicated genotype. Similar results were obtained in other two independent experiments. For analysis of leaf transmittance changes, detached leaves (from 16 day-old seedlings) were placed on 1% gellan gum medium in a 96-well plate, and the red light (650 nm) transmittance was automatically recorded every 2 min using a microplate reader (Versa-Max, Molecular Devices). Blue light was provided from a blue light-emitting diode (LED) illuminator (LED-mB, EYELA, Japan). The light conditions are shown at the top of the graph. D, Dark; BL, blue light. The decrease in the transmittance induced by 3 µmolm−2s−1 of blue light represents the chloroplast accumulation response, whereas the increase in the transmittance induced by 20 and 50 µmolm−2s−1 of blue light represents the avoidance response.
Collectively, our structural and functional analyses indicate that the JAC J-domain has structural features necessary for the Hsc70 activation, and suggest that the cooperation between JAC1 and Hsc70 is indispensable for chloroplast photorelocation movement. Among the fourteen Hsp70 family members,25 five cytosolic Hsc70s, Hsc70-1 to Hsc70-5, are candidates for the JAC1 partner, since JAC1 is localized in the cytosol.15 We obtained T-DNA knockout plants for Hsc70-1 (SALK_135531,26–28), Hsc70-2 (SALK_085076,26,27 and SALK_087844,28) and Hsc70-5 (SAIL_194_G12), and constructed hsc70-1/hsc70-5 and hsc70-2/hsc70-5 double mutants. Note that Hsc70-1 and Hsc70-2 are neighboring genes and thus the generation of hsc70-1/hsc70-2 double mutant by genetic crossing is not realistic. We found that these double knockout lines showed normal phenotype of chloroplast photorelocation movement (Suetsugu N, Wada M, unpublished data). These results are not unexpected, because the functional redundancy precludes the functional analyses of the Hsc70 genes.26–28 The identification of the in vivo Hsp70 partner(s) of the JAC1 J-domain will shed light on the understanding of JAC1 functions in chloroplast photorelocation movement.
The Possibility that JAC1 Mediates Chloroplast Photorelocation Movement via Clathrin-dependent Endocytosis
The structural similarity between the JAC1 and auxilin J-domains provides an open question: Is clathrin-dependent endocytosis involved in chloroplast photorelocation movement? A JAC1- overexpressed line, but not jac1 null mutant, showed decreased endocytosis activity in root hair cells compared to those of wild type.29 Recently, accumulating evidence indicates that the endocytosis and trafficking of membrane receptors are essential for plant development and physiological responses.30 Both phot1 and phot2 are internalized into the cytosol in a blue-light dependent manner.31–34 Importantly, both phototropins interacted to clathrin heavy chains in vivo, and the blue light-induced internalization of phot1 was inhibited by the treatment with Tyrphostin A23, an inhibitor of the clathrin-mediated endocytosis.34 Furthermore, yeast twohybrid screening for phot1-interacting proteins identified two ADP-ribosylation factors (ARFA1e and ARFB1b), GTPase members involved in clathrin-mediated endocytosis.35 Blue or white light illumination, but not red light, abrogated the interaction between phot1 and these ARFs in yeast, implicating the signaling role of ARFs in the phototropin-mediated blue light signaling pathway.35 Since yeast auxilin Swa2p was identified as a factor for the ARF-dependent protein transport pathways,36 it is plausible that JAC1 may cooperate with ARF for the phototropin endocytosis. The jac1 mutant showed normal phototropin protein expression, normal phototropin-mediated responses (leaf expansion and phototropism) and normal phot2-mediated chloroplast avoidance response, suggesting that jac1 mutant does not completely lack the phototropin functions. Experiment on blue light-induced internalization of phototropins in the jac1 mutant is required to elucidate the function of JAC1 in clathrin-mediated endocytosis.
Acknowledgements
SALK and SAIL T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center. This work was supported in part by the Japanese Ministry of Education, Sports, Science and Technology (MEXT 13139203 and 17084006 to M.W.; Targeted Proteins Research Program to D.K. and A.T.) and the Japan Society of Promotion of Science (JSPS 13304061, 16107002 and 20227001 to M.W.; 20870030 to N.S.). This work was partly performed in the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University.
Abbreviations
- CHUP1
chloroplast unusual positioning 1
- Hsc70
heat shock protein cognate 70
- JAC1
J-domain protein required for chloroplast accumulation response 1
- KAC1
kinesin-like protein for actin-based chloroplast movement 1
- phot
phototropin
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13915
References
- 1.Suetsugu N, Wada M. Chloroplast photorelocation movement mediated by phototropin family proteins in green plants. Biol Chem. 2007;388:927–935. doi: 10.1515/BC.2007.118. [DOI] [PubMed] [Google Scholar]
- 2.Suetsugu N, Wada M. Chloroplast photorelocation movement. In: Sandelius AS, Aronsson H, editors. The Chloroplasts. Plant Cell Monographs Series. Springer Berlin: Heiderberg; 2009. pp. 235–266. [Google Scholar]
- 3.Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M. Chloroplast avoidance movement reduces photodamage in plants. Nature. 2002;420:829–832. doi: 10.1038/nature01213. [DOI] [PubMed] [Google Scholar]
- 4.Suetsugu N, Dolja VV, Wada M. Why have chloroplasts developed a unique motility system? Plant Signal Behav. 2010;5:1190–1196. doi: 10.4161/psb.5.10.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, et al. Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, et al. Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 7.Jarillo JA, Gabryś H, Capel J, Alonso JM, Ecker JR, Cashmore AR. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- 8.DeBlasio SL, Luesse DL, Hangarter RP. A plant-specific protein essential for blue-light-induced chloroplast movements. Plant Physiol. 2005;139:101–114. doi: 10.1104/pp.105.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luesse DL, DeBlasio SL, Hangarter RP. Plastid movement impaired 2, a new gene involved in normal blue-light-induced chloroplast movements in Arabidopsis. Plant Physiol. 2006;141:1328–1337. doi: 10.1104/pp.106.080333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, et al. CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell. 2003;15:2805–2815. doi: 10.1105/tpc.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oikawa K, Yamasato A, Kong SG, Kasahara M, Nakai M, Takahashi F, et al. Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 2008;148:829–842. doi: 10.1104/pp.108.123075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt von Braun S, Schleiff E. The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta. 2008;227:1151–1159. doi: 10.1007/s00425-007-0688-7. [DOI] [PubMed] [Google Scholar]
- 13.Suetsugu N, Yamada N, Kagawa T, Yonekura H, Uyeda TQP, Kadota A, et al. Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:8860–8865. doi: 10.1073/pnas.0912773107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadota A, Yamada N, Suetsugu N, Hirose M, Saito C, Shoda K, et al. Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:13106–13111. doi: 10.1073/pnas.0906250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suetsugu N, Kagawa T, Wada M. An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 2005;139:151–162. doi: 10.1104/pp.105.067371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 17.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh P, Bursać D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenberg E, Greene LE. Multiple roles of auxilin and Hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 20.Takano A, Suetsugu N, Wada M, Kohda D. Crystallographic and functional analyses of J-domain of JAC1 essential for chloroplast photorelocation movement in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1372–1376. doi: 10.1093/pcp/pcq089. [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Taylor AB, Prasad K, Ishikawa-Brush Y, Hart PJ, Lafer EM, et al. Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface. Biochemistry. 2003;42:5748–5753. doi: 10.1021/bi034270g. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, et al. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutkiewicz R, Schilke B, Cheng S, Knieszner H, Craig EA, Marszalek J. Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. J Biol Chem. 2004;279:29167–29174. doi: 10.1074/jbc.M402947200. [DOI] [PubMed] [Google Scholar]
- 24.Xiao J, Kim LS, Graham TR. Dissection of Swa2p/auxilin domain requirements for cochaperoning Hsp70 clathrin-uncoating activity in vivo. Mol Biol Cell. 2006;17:3281–3290. doi: 10.1091/mbc.E06-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, et al. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6:201–208. doi: 10.1379/1466-1268(2001)006<0201:gaoths>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noël LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S, Witte CP, et al. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell. 2007;19:4061–4076. doi: 10.1105/tpc.107.051896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cazalé AC, Clément M, Chiarenza S, Roncato MA, Pochon N, Creff A, et al. Altered expression of cytosolic/nuclear HSC70-1 molecular chaperone affects development and abiotic stress tolerance in Arabidopsis thaliana. J Exp Bot. 2009;60:2653–2664. doi: 10.1093/jxb/erp109. [DOI] [PubMed] [Google Scholar]
- 28.Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, et al. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell. 2009;21:3984–4001. doi: 10.1105/tpc.109.071548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezaki B, Kiyohara H, Matsumoto H, Nakashima S. Overexpression of an auxilin-like gene (F9E10.5) can suppress Al uptake in roots of Arabidopsis. J Exp Bot. 2007;58:497–506. doi: 10.1093/jxb/erl221. [DOI] [PubMed] [Google Scholar]
- 30.Geldner N, Robatzek S. Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 2008;147:1565–1574. doi: 10.1104/pp.108.120287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto K, Briggs WR. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong SG, Suzuki T, Tamura K, Mochizuki N, Hara-Nishimura I, Nagatani A. Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 2006;45:994–1005. doi: 10.1111/j.1365-313X.2006.02667.x. [DOI] [PubMed] [Google Scholar]
- 33.Wan YL, Eisinger W, Ehrhardt D, Kubitscheck U, Baluska F, Briggs WR. The subcellular localization and blue-light-induced movement of phototropin 1-GFP in etiolated seedlings of Arabidopsis thaliana. Mol Plant. 2008;1:103–117. doi: 10.1093/mp/ssm011. [DOI] [PubMed] [Google Scholar]
- 34.Kaiserli E, Sullivan S, Jones MA, Feeney KA, Christie JM. Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. Plant Cell. 2009;21:3226–3244. doi: 10.1105/tpc.109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan S, Thomson CE, Kaiserli E, Christie JM. Interaction specificity of Arabidopsis 14-3-3 proteins with phototropin receptor kinases. FEBS Lett. 2009;583:2187–2193. doi: 10.1016/j.febslet.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Gall WE, Higginbotham MA, Chen CY, Ingram MF, Cyr DM, Graham TR. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr Biol. 2000;10:1349–1358. doi: 10.1016/s0960-9822(00)00771-5. [DOI] [PubMed] [Google Scholar]