Abstract
Plants respond to developmental cues and environmental stresses by controlling both the level and activity of various hormones. One mechanism of modulating hormone action involves amino acid conjugation. In plants, the GH3 family of enzymes conjugates various amino acids to jasmonates, auxins and benzoates. The effect of conjugation can lead to activation, inactivation or degradation of these molecules. Although the acyl acid and amino acid specificities of a few GH3 enzymes have been examined qualitatively, further in-depth analysis of the structure and function of these proteins is needed to reveal the molecular basis for how GH3 proteins modulate plant hormone action.
Key words: GH3 protein, auxin, jasmonate, plant hormone regulation, adenylation reaction
Plant hormones control the interplay of a host of growth and developmental processes. Although the exact mechanisms of plant hormone sensing and signal response vary widely,1–5 the auxin and jasmonate hormone response systems share similar mechanisms of action. Auxins, primarily indole-3-acetic acid (IAA), are involved in cell division, elongation and differentiation. The jasmonates (JA) play roles in seed germination, fertility, root growth and pathogen responses.1–3 Perception of IAA and JA by either the SCFTIR1 ubiquitin ligase complex6–9 or the SCFCOI1 ubiquitin ligase complex,10–12 respectively, recruits different classes of transcriptional modulators (i.e., Aux/IAA proteins and JAZ proteins, respectively) to the complex for ubiquitination. Degradation of Aux/IAA proteins modulates expression of auxin-responsive genes and degradation of JAZ proteins alters transcription of jasmonate-responsive genes. A critical component in these plant hormone responses is the pre-receptor action of various enzyme systems that maintain both the levels and forms of small molecule signals.
Changes in cellular hormone concentrations and the chemical structure of a hormone will affect interaction with cognate receptors and alter developmental processes and responses. Biochemically, the levels and molecular structure of plant hormones are modified by the rates of their biosynthesis, catabolism, and/or modification. For example, in Arabidopsis thaliana, less than 5% of auxins are found in the active free form with the remaining 95% primarily conjugated to either amino acids or peptides.13 Enzymatic conversion of unconjugated JA and IAA into the corresponding amino acid conjugates drastically alters hormone action (Fig. 1). Addition of isoleucine to JA results in synthesis of the bioactive jasmonate hormone (JA-isoleucine) that binds to the COI1 receptor to trigger JA-mediated responses.14 In contrast, formation of IAA-aspartate and IAA-glutamate marks auxin for subsequent metabolic degradation and conjugation of IAA with either alanine or leucine maintains a ready pool for formation of the active free acid form of auxin.15,16 Thus, metabolism of IAA and JA provides a simple means of modulating plant hormone receptor occupancy and controlling gene responses. Some GH3 family of proteins catalyze the conjugations of various amino acids to plant hormones, including JA and IAA; however, a majority of GH3 proteins in plants remains uncharacterized.
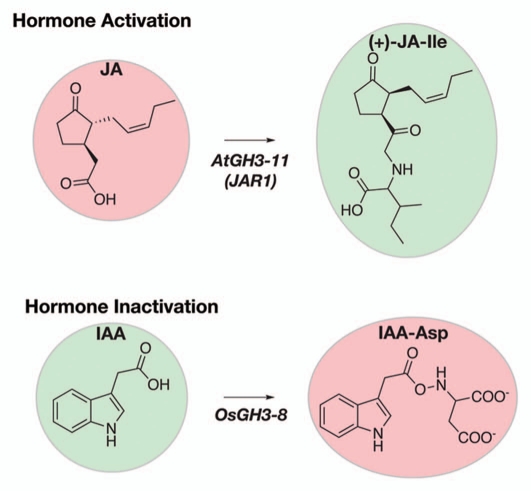
Figure 1.
Pre-Receptor Enzyme Action by GH3 Enzymes. Modification of jasmonic acid (JA) and indole-3-acetic acid (IAA; auxin) directly effects plant hormone potency. Conjugation of JA with isoleucine by AtGH3-11 leads to formation of the active jasmonate hormone (+)-JA-Ile (top part). Conjugation of IAA with aspartate by OsGH3-8 yields IAA-Asp, which leads to degradation of the active hormone (bottom part). Red and green correspond to inactive and active hormone forms, respectively.
The GH3 Family: A Common Gene Family in Plants
The first GH3 gene was identified in 1984 as an early auxin-responsive gene in Glycine max (soybean).17 Since then, GH3-related genes have been found in multiple plant species; however, not all of these genes are auxin-responsive. In the model dicot species Arabidopsis thaliana (thale cress), there are 19 GH3 genes.18 GH3 transcripts have also been reported in tobacco, pungent pepper and tomato.18–22 In the genome of the model monocot Oryza sativa (rice) genome, there are 13 GH3 genes.23 Transcripts have been detected, but not characterized, in other monocots, including wheat, maize, sorghum and barley.18,23 In addition, GH3 genes are found in the gymnosperm Pinus pinaster and the moss Physomitrella patens.24–26 EST analysis suggests that GH3 genes are found across multiple plants, mosses and algae.18,23 Phylogenetic analysis of the GH3 families in Arabidopsis and rice show that these sequences fall into three distinct groups (I–III), although rice lacks group III members,18,23 as summarized in Table 1. To date, a number of GH3 genes from Arabidopsis and rice have been cloned, mainly through morphological screening of either loss-of- or gain-of-function mutants.
Table 1.
Summary of GH3 family members in Arabidopsis and Rice
| GH3 protein | Groupa | Mutantb | Acyl acidc | Amino acidc | Refs |
| AtGH3-1 | II | ? | ? | 22 | |
| AtGH3-2 | II | ydk1 | IAA | Met, Trp | 22, 29 |
| AtGH3-3 | II | IAA | Asp, Met, Try, Trp | 22 | |
| AtGH3-4 | II | IAA | Asp, Met, Trp | 22 | |
| AtGH3-5 | II | wes1 | IAA | Glu, Asp, Met, Tyr, Trp | 14, 22, 30–34 |
| AtGH3-6 | II | dfl1 | IAA | Asp, Met, Trp | 22, 35 |
| AtGH3-7 | III | ? | ? | ||
| AtGH3-8 | III | ? | ? | ||
| AtGH3-9 | II | ? | ? | 36–37 | |
| AtGH3-10 | I | dfl2 | ? | ? | 28 |
| AtGH3-11 | I | jar1/fin219 | JA | Ile | 14, 27, 46 |
| AtGH3-12 | III | pbs3/gdg1 | benzoates | Glu, His, Leu, Met | 43–45 |
| AtGH3-13 | III | ? | ? | ||
| AtGH3-14 | III | ? | ? | ||
| AtGH3-15 | III | ? | ? | ||
| AtGH3-16 | III | ? | ? | ||
| AtGH3-17 | II | IAA | Glu, Met, Trp | 22 | |
| AtGH3-18 | III | ? | ? | ||
| AtGH3-19 | III | ? | ? | ||
| OsGH3-1 | II | ? | ? | 41 | |
| OsGH3-2 | II | ? | ? | ||
| OsGH3-3 | I | ? | ? | ||
| OsGH3-4 | II | ? | ? | ||
| OsGH3-5 | I | ? | ? | ||
| OsGH3-6 | I | ? | ? | ||
| OsGH3-7 | II | ? | ? | ||
| OsGH3-8 | II | IAA | Asp | 38–40 | |
| OsGH3-9 | II | ? | ? | ||
| OsGH3-10 | II | ? | ? | ||
| OsGH3-11 | II | ? | ? | ||
| OsGH3-12 | I | ? | ? | ||
| OsGH3-13 | II | tld1 | IAA | Glu, Asp, Phe, Leu, Ala | 42 |
Mutant associated with GH3 gene.
Major acyl acid and amino acid substrates of the GH3 protein identified by substrate screening assays. Substrates in bold have been confirmed by steady-state kinetic analysis of purified protein. Question marks indicate unknown enzymatic activity and substrate specificity.
One of the two group I proteins in Arabidopsis is directly linked to JA hormone action. The A. thaliana GH3-11 (AtGH3-11) protein was first characterized by the Staswick group after identifying the jar1 mutant, which showed insensitivity to JA treatment.14 AtGH3-11 is essential for formation of the bioactive jasmonate JA-isoleucine14 (Fig. 1). A different allele of this gene (i.e., fin219) was previously isolated from a screen to identify extragenic modifier mutations of cop1, a temperature-sensitive allele of the photomorphogenic development repressor COP1; however, no activity was assigned to the encoded protein.27 The other group I GH3 in Arabidopsis, AtGH3-10, is associated with the dfl2 mutant and is involved in red light-specific hypocotyl elongation, but no enzymatic activity has been reported for this protein.28
Characterization of multiple group II GH3 genes in Arabidopsis also demonstrates critical roles in plant growth and development primarily through the conjugation of IAA to amino acids. For AtGH3-2, a gain-of-function mutant ydk1-D was found through a short primary root, reduced lateral root number and reduced apical dominance phenotype.29 AtGH3-5 appears to be involved in both IAA and salicylic acid (SA) action.14,30–31 AtGH3-5 was first characterized in a promoter trap line that exhibited light-dependent reporter gene expression32 and later by the loss-of-function mutant wes1 and two gain-of-function mutants wes1-D and gh3.5-1D.14,33,34 AtGH3-6 was isolated as the dfl1 mutant, which negatively regulates shoot elongation and lateral root formation, but positively regulates the light response of hypocotyl length.35 Later biochemical analysis of AtGH3-2, -5, -6 showed that these enzymes conjugate IAA to various amino acids.22 In contrast to most group II GH3, AtGH3-9 expression is repressed by low concentrations of IAA and a loss-of-function mutant shows growth of longer primary roots.36,37
Three group II GH3 have been characterized in rice. The O. sativa GH3-8 (OsGH3-8) gene is auxin-responsive and functions in auxin-dependent development.38 Overexpression of OsGH3-8 shows abnormal morphology, but enhanced resistance to the rice pathogen Xanthomonas oryzae.38 The OsGH3-8 enzyme specifically conjugates IAA and aspartate to form IAA-aspartate.38–40 Less is known about the other two identified rice GH3 genes. Overexpression of OsGH3-1 in rice causes dwarfism, significantly reduces free auxin levels and cell elongation, but increases resistance to fungal pathogens.41 Similarly, a gain-of-function mutant, tld1-D, in the OsGH3-13 gene results in increased tillers, enlarged leaf angles, dwarfism and improved drought tolerance.42
The only characterized group III GH3 member is AtGH3-12.43–45 Three pbs3 mutants display increased disease susceptibility to virulent and avirulent Pseudomonas syringae.43 The gdg1 mutant is allelic to pbs3.45
GH3 Proteins: Acyl Acid Amido Synthetases
Although the GH3 proteins are wide-spread across the plant kingdom, relatively little is known about their metabolic roles and kinetic properties. The first insight on the biochemical function of the GH3 proteins was provided by analysis of the group I AtGH3-11 or JAR1, protein.14 Sequence comparisons detected low amino acid similarity between AtGH3.11 and the firefly luciferase superfamily of adenylating enzymes, suggesting a possible overall reaction for the GH3 proteins.14 Adenylating enzymes catalyze the ATP-dependent activation of carboxyl groups on a variety of substrates by formation of an adenylated reaction intermediate.46 Formation of the activated intermediate allows for subsequent reactions with other substrates.46 Initial work on AtGH3-11 (JAR1) demonstrated that it catalyzes the formation of JA-Ile.47 Subsequent studies of the Arabidopsis group II AtGH3-2, -3, -4, -5, -6 and -17 show that these enzymes accept IAA as a substrate to function as acyl acid amido synthetases.22 Based on screening of amino acid preference using a thin-layer chromatography (TLC)-based assay, it was suggested that these GH3 proteins utilize a broad range of amino acid subtrates,22 as summarized in Table 1. Using a liquid chromatography-mass spectrometry (LC-MS) assay, OsGH3-13 also conjugates multiple amino acids to IAA.42 The highest activity of OsGH3-13 was observed with glutamate, but aspartate, phenylalanine, leucine and alanine are also substrates for the enzyme.42 Surprisingly, the kinetic parameters for only two GH3 proteins (OsGH3-8,39,40 and AtGH3-12,45) have been determined to date.
Recently, we used OsGH3-8, which specifically functions as an IAA-aspartate amido synthetase, for analysis of the chemical and kinetic mechanism of a GH3 protein.39,40 These studies demonstrate formation of a chemically competent adenylation intermediate in the reaction sequence and define the order of substrate/product binding and release.40 Using both a MS-based assay that directly monitored product formation and a spectrophotometric assay coupled to AMP release following IAA-Asp formation, the kinetic parameters for the overall reaction catalyzed by OsGH3-8 were determined.39,40 Each assay yields comparable kinetic values,39,40 as summarized in Table 2. Importantly, these assays provide a quantitative assessment of GH3 protein acyl acid and amino acid specificities. OsGH3-8 is highly specific for IAA (150 nmolmin−1mgprotein−1) compared to JA (0.3 nmol min−1 mg protein−1) and SA (1.1 nmol min−1 mg protein−1). Other auxin-analogs, including phenylacetic acid, indole butyric acid and napthanlenacid acid, were also substrates with catalytic efficiencies (i.e., kcat/Km) 1.4- to 9-fold lower than that observed for IAA.40 Similarly, the enzyme is also highly specific for aspartate. Asparagine, the next best amino acid substrate, shows a 45-fold reduction in kcat/Km compared to aspartate and other amino acids are more than 100-fold lower.40 These results suggest that under biological conditions, it is highly unlikely that OsGH3-8 accepts amino acids other than aspartate as a substrate.
Table 2.
Comparison of kinetic parameters of AtGH3-12, OsGH3-8, and AtGH3-17
| Assay method | Kinetic parameter | Assay method | Kinetic parameter | |
| AtGH3-12 | HPLCa | Vmax = 20.4 min−1 | coupled UV/Visb | Vmax = 13.2 min−1 |
| HPLCa | Km4-HBA = 450 µM | coupled UV/Visb | Km4-HBA = 63 µM | |
| adenylationa | KmATP = 791 µM | coupled UV/Visb | KmATP = 99 µM | |
| none | not determined | coupled UV/Visb | KmGlu = 3.83 mM | |
| OsGH3-8 | mass specc | Vmax = 21.1 min−1 | coupled UV/Visd | Vmax = 20.6 min−1 |
| mass specc | KmIAA = 123 µM | coupled UV/Visd | KmIAA = 187 µM | |
| mass specc | KmATP = 50 µM | coupled UV/Visd | KmATP = 36 µM | |
| mass specc | KmAsp = 1.58 mM | coupled UV/Visd | KmAsp = 3.91 mM | |
| AtGH3-17 | coupled UV/Visb | Vmax = 7.4 min−1 | ||
| coupled UV/Visb | KmIAA = 58 µM | |||
| coupled UV/Visb | KmATP = 138 µM | |||
| coupled UV/Visb | KmGlu = 0.54 mM |
AtGH3-12 is the only biochemically studied group III enzyme. It favors 4-substituted benzoates, including 4-aminobenzoate and 4-hydroxybenzoate (4-HBA), as substrates, but accepts benzoate with moderate activity and is inactive with 2-substituted benzoates.45 In this study, the authors used various assays for activity analysis, including a spectrophotometric adenylation assay, which monitors only the first half of the chemical reaction sequence, to determine the Km value for ATP and an HPLC assay to determine Vmax and the Km for product formation using 4-HBA as a substrate (Table 2). Although no kinetic information for amino acid specificity was provided, a TLC assay was used to show that 4-HBA was conjugated to various amino acids. These results suggest that glutamate, histidine, lysine and methionine are the major substrates with isoleucine, leucine, valine, threonine, threonine, serine and tryptophan as minor substrates.
Studies on the group II AtGH3, AtGH3-12 and OsGH3-8 raise the question—are these proteins promiscuous or specific for amino acid substrates? Addressing this point, will help to define the possible biological roles of these proteins. For comparison, the OsGH3-8 spectrophotometric assay was used to re-evaluate AtGH3-12 and to determine the kinetic parameters for AtGH3-17 (Table 2). For AtGH3-12, the Vmax and Km values for 4-HBA, ATP and glutamate were re-determined and are comparable to those originally reported for this protein and to OsGH3-8. Importantly, AtGH3-12 shows an 80-fold preference for glutamate compared to the next best amino acid substrate, indicating that this enzyme is highly specific and unlikely to accept other amino acid substrates in vivo (Fig. 2). Analysis of AtGH3-17 shows that this enzyme displays kinetics similar to OsGH3-8 and AtGH3-12 (Table 2). In comparison to earlier studies, these experiments emphasize the need to use assays that accurately determine substrate specificity for further studies on the possible biochemical functions of the GH3 proteins in plants.
Figure 2.
Amino Acid Specificity of AtGH3-12. Purified protein was assayed by coupling AMP formation to a UV/Vis assay, as previously described in reference 40. Substrate concentrations were 1 mM IAA, 1 mM ATP and 5 mM amino acid. Data is shown as a mean ± SE. (n = 3).
Summary
Plant development, growth and fitness are all determined by the complex integration of multiple signaling pathways and hormone responses. As part of the system that regulates plant hormone effects, GH3 enzymes play important, but ill defined, roles, as evidenced by the various mutant phenotypes associated with these genes. The GH3 proteins appear to control levels of major plant hormones, including IAA and JA, and modulate pathways responsible for plant growth and development, seed development, light signaling, drought response, thermogenesis and pathogen resistance.13–45 Biochemically, the GH3 proteins are defined by their ability to catalyze the conjugation of acyl acids to amino acids via an adenylation reaction. Through a simple chemical modification, GH3 proteins can regulate levels of inactive and active forms of JA and IAA. For example, the ability of a GH3 protein to modify the chemical structure of multiple JA- and IAA-related substrates may allow for changes of metabolic pools across a biosynthetic pathway, in addition to modifying the final hormone product. Although initial work has qualitatively determined the substrate activity for a handful of GH3 proteins, further detailed analysis of the acyl acid and amino acid specificities of the GH3 family is an essential step toward defining the possible biological function of these enzymes. In time, complete analysis of the temporal and spatial expression patterns, cellular localization and specificity of enzymatic activities will provide insight on how this diverse protein family modulates plant hormone function.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13941
References
- 1.Santner A, Calderon-Villalobos LIA, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nature Chem Biol. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- 2.Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutler SR, Rodriguez PL, Finkelsteine RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Bio. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 5.Kim TW, Wang ZY. Brassinosteriod signal transduction from receptor kinases to transcription factors. Annu Rev Plant Bio. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 6.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 7.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 8.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 9.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 10.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 12.Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, et al. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol. 2002;50:309–332. doi: 10.1023/a:1016024017872. [DOI] [PubMed] [Google Scholar]
- 14.Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem. 2002;277:20446–20452. doi: 10.1074/jbc.M111955200. [DOI] [PubMed] [Google Scholar]
- 16.Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004;135:978–988. doi: 10.1104/pp.104.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagen G, Kleinschmidt A, Guilfoyle T. Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta. 1984;162:147–153. doi: 10.1007/BF00410211. [DOI] [PubMed] [Google Scholar]
- 18.Terol J, Domingo C, Talon M. The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene. 2006;371:279–290. doi: 10.1016/j.gene.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Roux C, Perrot-Rechenmann C. Isolation by differential display and characterization of a tobacco auxin-responsive cDNA Nt-gh3 related to GH3. FEBS Lett. 1997;419:131–136. doi: 10.1016/s0014-5793(97)01447-6. [DOI] [PubMed] [Google Scholar]
- 20.Lahey KA, Yuan R, Burns JK, Ueng PP, Timmer LW, Kuang-Ren C. Induction of phytohormones and differential gene expression in citrus flower infected by the fungus Colletotrichum acutatum. Mol Plant Microbe Interact. 2004;17:1394–1401. doi: 10.1094/MPMI.2004.17.12.1394. [DOI] [PubMed] [Google Scholar]
- 21.Liu K, Kang BC, Jiang H, Moore SL, Li H, Watkins CB, et al. A GH3-like gene, CcGH3, isolated from Capsicum chinense L. fruit is regulated by auxin and ethylene. Plant Mol Biol. 2005;58:447–464. doi: 10.1007/s11103-005-6505-4. [DOI] [PubMed] [Google Scholar]
- 22.Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain M, Kaur N, Tyagi AK, Khurana JP. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa) Funct Integr Genomics. 2006;6:36–46. doi: 10.1007/s10142-005-0142-5. [DOI] [PubMed] [Google Scholar]
- 24.Reddy SM, Hitchin S, Melayah D, Pandey AK, Raffier C, Henderson J, et al. The auxin-inducible GH3 homologue Pp-GH3.16 is downregulated in Pinus pinaster root systems on ectomycorrhizal symbiosis establishment. New Phytol. 2006;17:391–400. doi: 10.1111/j.1469-8137.2006.01677.x. [DOI] [PubMed] [Google Scholar]
- 25.Bierfreund NM, Tintelnot S, Reski R, Decker EL. Loss of GH3 function does not affect phytochrome-mediated development in a moss, Physcomitrella patens. J Plant Physiol. 2004;161:823–835. doi: 10.1016/j.jplph.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig-Müller J, Jülke S, Bierfreund NM, Decker EL, Reski R. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 2009;181:323–338. doi: 10.1111/j.1469-8137.2008.02677.x. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, et al. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- 28.Takase T, Nakazawa M, Ishikawa A, Manabe K, Matsui M. DFL2, a new member of the Arabidopsis GH3 gene family, is involved in red light-specific hypocotyl elongation. Plant Cell Physiol. 2003;44:1071–1080. doi: 10.1093/pcp/pcg130. [DOI] [PubMed] [Google Scholar]
- 29.Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, et al. Ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004;37:471–483. doi: 10.1046/j.1365-313x.2003.01973.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, et al. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007;145:450–464. doi: 10.1104/pp.107.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Wang M, Li Z, Li Q, He Z. Arabidopsis GH3.5 regulates salicylic acid-dependent and both NPR1-dependent and independent defense responses. Plant Signal Behav. 2008;3:537–542. doi: 10.4161/psb.3.8.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka S, Mochizuki N, Nagatani A. Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol. 2002;43:281–289. doi: 10.1093/pcp/pcf033. [DOI] [PubMed] [Google Scholar]
- 33.Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, et al. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem. 2007;282:10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- 34.Park JE, Seo PJ, Lee AK, Jung JH, Kim YS, Park CM. An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol. 2007;48:1236–1241. doi: 10.1093/pcp/pcm086. [DOI] [PubMed] [Google Scholar]
- 35.Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, et al. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation and positively regulates the light response of hypocotyl length. Plant J. 2001;25:213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- 36.Khan S, Stone JM. Arabidopsis thaliana GH3.9 influences primary root growth. Planta. 2007;226:21–34. doi: 10.1007/s00425-006-0462-2. [DOI] [PubMed] [Google Scholar]
- 37.Khan S, Stone JM. Arabidopsis thaliana GH3.9 in auxin and jasmonate cross talk. Plant Signal Behav. 2007;2:483–485. doi: 10.4161/psb.2.6.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, et al. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008;20:228–240. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Q, Zhang B, Hicks LM, Wang S, Jez JM. A liquid chromatography-tandem mass spectrometry-based assay for indole-3-acetic acid-amido synthetase. Anal Biochem. 2009;390:149–154. doi: 10.1016/j.ab.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q, Westfall CS, Hicks LM, Wang S, Jez JM. Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J Biol Chem. 2010;285:29780–29786. doi: 10.1074/jbc.M110.146431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domingo C, Andrés F, Tharreau D, Iglesias DJ, Talón M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant Microbe Interact. 2009;22:201–210. doi: 10.1094/MPMI-22-2-0201. [DOI] [PubMed] [Google Scholar]
- 42.Zhang SW, Li CH, Cao J, Zhang YC, Zhang SQ, Xia YF, et al. Altered architecture and enhanced drought tolerance in rice via the downregulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009;151:1889–1901. doi: 10.1104/pp.109.146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jagadeeswaran G, Raina S, Acharya BR, Maqbool SB, Mosher SL, Appel HM, et al. Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 2007;51:234–246. doi: 10.1111/j.1365-313X.2007.03130.x. [DOI] [PubMed] [Google Scholar]
- 44.Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, et al. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007;144:1144–1156. doi: 10.1104/pp.107.097691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okrent RA, Brooks MD, Wildermuth MC. Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J Biol Chem. 2009;284:9742–9754. doi: 10.1074/jbc.M806662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gulick AM. Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases and firefly luciferase. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guranowski A, Miersch O, Staswick PE, Suza W, Wasternack C. Substrate specificity and products of side-reactions catalyzed by jasmonate: amino acid synthetase (JAR1) FEBS Lett. 2007;581:815–820. doi: 10.1016/j.febslet.2007.01.049. [DOI] [PubMed] [Google Scholar]