Abstract
Effector proteins expressed in the esophageal gland cells of cyst nematodes are delivered into plant cells through a hollow, protrusible stylet. Although evidence indicates that effector proteins function in the cytoplasm of the syncytium,1–3 technical constraints have made it difficult to directly determine where nematode effector proteins are initially delivered: cytoplasm, extracellular space, or both. Recently, we demonstrated that soybean cyst nematode CLE (HgCLE) propeptides are delivered to the cytoplasm of syncytial cells. Genetic and biochemical analyses indicate that the variable domain (VD) sequence is then required for targeting cytoplasmically delivered nematode CLEs to the apoplast where they function as ligand mimics of endogenous plant CLE peptides.4 The fact that nematode CLEs are targeted through the gland cell secretory pathway and delivered as mature propeptides into plant cells makes it impossible for these proteins to be subsequently delivered to the extracellular space via co-translational translocation through the endoplasmic reticulum (ER) secretory pathway of the host cell. However, when expressed in transgenic plants, if the mature nematode CLE propeptide harbored a functional cryptic signal peptide, it could possibly traffic to the apoplast through the ER secretory pathway by co-translational translocation. Here, we present evidence that VDI, the N-terminal sequence of the VD of HgCLE2,4 is sufficient for trafficking CLE peptides to the apoplast and that trafficking is indeed through an alternative pathway other than co-translational translocation.
Key words: cyst nematode, effector, CLE, variable domain, trafficking, endoplasmic reticulum, co-translational translocation, post-translational
VDI is Sufficient to Target CLE Peptides to the Extracellular Space
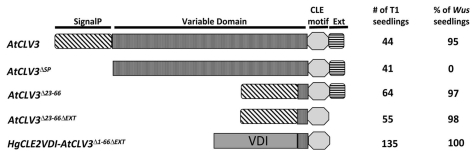
All plant CLE proteins contain N-terminal signal peptides that can target plant CLEs through the ER secretory pathway to function in the extracellular space.5,6 Overexpression of Arabidopsis CLAVATA3 (AtCLV3),7 the best studied plant CLE gene, suppresses the expression of the homeobox gene WUSCHEL (WUS),8 causing a wus mutant phenotype. Overexpression of AtCLV3ΔSP abolishes its function because AtCLV3 is not properly targeted to the extracellular space, where it has been shown to function4,6 (Fig. 1). The same has recently been shown for several other Arabidopsis CLEs.9 In a previous study of AtCLV3,10 deletion of the VD (AtCLV3Δ23–66) or of the extension sequence downstream of the AtCLV3 CLE motif (AtCLV3ΔEXT) did not affect function. However, four amino acids of the VD immediately N-terminal of the 12-amino acid CLE motif were included in the VD deletion construct. In this study, deletion of both the VD and the extension sequence (AtCLV3Δ23–66ΔEXT) did not affect AtCLV3 function (Fig. 1), which indicates that 16 amino acids (amino acids 67–82) spanning the CLE motif are sufficient for AtCLV3 function when targeted to the apoplast by its signal peptide. Recently, we demonstrated that the nematode HgCLE2 VD can target the CLE motif of HgCLE2 and AtCLV3 to the apoplast for function.4 In this study, we tested whether the N-terminal sequence of the VD (VDI) is sufficient for the trafficking function. VDI was fused with the 16-aa AtCLV3 CLE motif region sequence and overexpressed in Arabidopsis. All 135 T1 seedlings displayed clear wus phenotypes (Fig. 1), which suggests that VDI of HgCLE2 is sufficient to target CLE peptides to the extracellular space for function. Since these results are based on overexpression studies in Arabidopsis, we speculated that VDI might harbor a cryptic signal peptide sequence involved in targeting CLE peptides to the ER secretory pathway to function in the extracellular space.
Figure 1.
Overexpression of HgCLE2 variable domain I (VDI) targets the AtCLV3 CLE domain to the apoplast for function in Arabidopsis. Design of overexpression constructs. AtCLV3, full length AtCLV3; AtCLV3ΔSP, deletion of signal peptide from AtCLV3; AtCLV3Δ23–66, deletion of variable domain (VD) from AtCLV3; AtCLV3Δ23–66ΔEXT, deletion of both the VD and extension sequence after the CLE motif from AtCLV3; HgCLE2VDI-AtCLV3Δ1–66ΔEXT, VDI of HgCLE2 fused to the 16-amino acid CLE motif region sequence of AtCLV3.
Identification of a Predicted Cryptic Signal Peptide Sequence in VDI
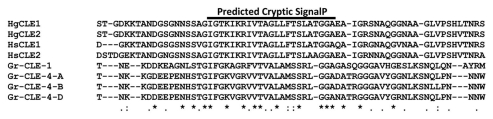
The presence of a putative cryptic signal peptide in the VDI sequence of HgCLE2 was suggested by a bioinformatics analysis. Two independent bioinformatics protocols were designed that employed the SignalP version 3.0 software.11 Both protocols predicted a cryptic signal peptide in VDI. In the first protocol, a signal peptide was detected using the entire sequence of HgCLE2. In the second protocol, an N-terminal region of HgCLE2 (residues 1–41) was removed, and signal peptide detection was repeated to confirm that the region does not affect the prediction score. This established the left boundary of the predicted peptide. To determine the right boundary, 5, 10 and 15 residues were incrementally removed from the remaining sequence and reanalyzed with SignalP using the same parameters. Together, the bioinformatics analysis predicted a cryptic signal peptide in VDI between residues I42 and A64 of HgCLE2 (Fig. 2).
Figure 2.
Sequence alignment of the variable domain I (VDI) region of different cyst nematode CLE proteins reveals a high level of sequence conservation. Hg, Heterodera glycines; Hs, Heterodera schachtii; Gr, Globodera rostochiensis.
Besides the two CLE proteins from soybean cyst nematode,4 we recently reported on the identification of two CLE gene sequences, HsCLE1 and HsCLE2, from the beet cyst nematode (Heterodera schachtii), a close relative of soybean cyst nematode.12 Several CLE sequences from potato cyst nematode (Globodera rostochiensis) have also been reported.13 So far, all of the cyst nematode CLEs identified to date can mimic plant CLEs when overexpressed in plants without their signal peptide sequences.4,12,13 These data indicate that the function of the VD in retargeting CLE peptides to the apoplast is conserved across cyst nematodes. Not surprisingly, a T-Coffee sequence alignment among nematode CLEs revealed a high level of sequence conservation in VDI spanning the region that contains the predicted cryptic signal peptide sequence in HgCLE2 (Fig. 2). This suggests that cyst nematodes may utilize the same mechanism to traffic CLE proteins to the extracellular space of plant cells.
VDI Directs Post-Translational Targeting of CLE Peptides to the Extracellular Space
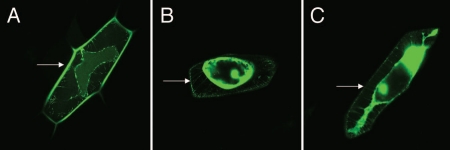
To test the function of the putative cryptic signal sequence, a C-terminal eGFP translational fusion to VDI was constructed to determine subcellular localization in onion epidermal cells. AtCLV3SP-eGFP and 35S-eGFP were included as positive and negative controls, respectively. These constructs were transiently expressed in onion epidermal cells by gold particle bombardment, and the GFP signal in the extracellular space was examined after plasmolysis.14 AtCLV3SP-eGFP effectively targeted eGFP to the extracellular space (Fig. 3A); whereas, cells expressing 35S-eGFP accumulated GFP in the cytoplasm and nucleus, not in the extracellular space (Fig. 3B). Cells expressing HgCLE2VDI-eGFP accumulated GFP in the cytoplasm and nucleus similar to 35S-eGFP (Fig. 3C). These data indicate that the identified cryptic signal peptide is not functioning as a canonical ER-targeting sequence; therefore, VDI targeting of nematode CLEs to the extracellular space by co-translational translocation into the ER secretory pathway does not explain the results of overexpression studies. This is consistent with the fact that the mature CLE propeptide is delivered into syncytial cells by the nematode. Thus, VDI is targeting nematode CLEs to the extracellular space post-translationally through an alternative trafficking mechanism, which may be precluded by the GFP tag in this assay because of its molecular weight and/or structural differences from CLE peptides.
Figure 3.
Localization of eGFP in onion epidermal cells after plasmolysis. (A) AtCLV3SP-eGFP; (B) 35S-eGFP; (C) HgCLE2VDI-eGFP. Arrows point to eGFP accumulation in the extracellular space in (A), but not in (B and C).
A BLAST search15 to identify proteins that contain a subsequence similar to the cryptic signal peptide identified in HgCLE2 VDI was performed. The BLAST search detected proteins that share a surprisingly long stretch of 9 identical residues or 10–11 similar residues when considering positive substitutions with the cryptic signal peptide. An alignment using the full-length HgCLE2 sequence with these proteins revealed only ∼3–7% sequence identity, suggesting that these proteins are not related. Interestingly, however, a large number of these proteins are transporters from a number of different species, including plants (e.g., Oryza sativa) and bacteria (e.g., Burkholderia vietnamiensis). Further studies will be necessary to determine exactly how the VDI sequence facilitates transport of nematode CLEs to the extracellular space.
Acknowledgements
This work was supported by the USDA-NRI Competitive Grants Program (grant nos 2007-35607-17790 and 2009-35302-05304) to M.G.M. a USDA Special Grant (grant no. 2008-34113-19420) to M.G.M. and an NSF CAREER Award (DBI-0845196) to D.K. We also thank Melody Kroll for assistance with editing.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13774
References
- 1.Elling AA, Davis EL, Hussey RS, Baum TJ. Active uptake of cyst nematode parasitism proteins into the plant cell nucleus. Int J Parasitol. 2007;37:1269–1279. doi: 10.1016/j.ijpara.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Hewezi T, Howe P, Maier TR, Hussey RS, Mitchum MG, Davis EL, et al. Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. Plant Cell. 2008;20:3080–3093. doi: 10.1105/tpc.108.063065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacco MA, Koropacka K, Grenier E, Jaubert MJ, Blanchard A, Goverse A, et al. The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2- and RanGAP2-dependent plant cell death. PLoS Pathog. 2009;5:1000564. doi: 10.1371/journal.ppat.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Lee C, Replogle A, Joshi S, Korkin D, Hussey RS, et al. Dual roles for the variable domain in protein trafficking and host-specific recognition of Heterodera glycines CLE effector proteins. New Phytol. 2010;187:1003–1017. doi: 10.1111/j.1469-8137.2010.03300.x. [DOI] [PubMed] [Google Scholar]
- 5.Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126:939–942. doi: 10.1104/pp.126.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell. 2002;14:969–977. doi: 10.1105/tpc.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 8.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 9.Meng L, Ruth KC, Fletcher JC, Feldman L. The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Mol Plant. 2010;3:760–772. doi: 10.1093/mp/ssq021. [DOI] [PubMed] [Google Scholar]
- 10.Fiers M, Golemiec E, van Der Schors R, van Der Geest L, Li KW, Stiekema WJ, et al. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 2006;141:1284–1292. doi: 10.1104/pp.106.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bendtsen JD, Nielsen H, Von Heijne G, Brunak G. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Replogle A, Hussey RS, Baum TJ, Wang X, Davis EL, et al. Identification of potential host plant mimics of CLV3/ESR(CLE)-like peptides from the plant-parasitic nematode Heterodera schachtii. Mol Plant Pathol. 2010:177–186. doi: 10.1111/J.1364-3703.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu SW, Chen S, Wang J, Yu H, Chronis D, Mitchum MG, et al. Structural and functional diversity of CLAVATA3/ESR (CLE)-like genes from the potato cyst nematode Globodera rostochiensis. Mol Plant Microbe Interact. 2009;22:1128–1142. doi: 10.1094/MPMI-22-9-1128. [DOI] [PubMed] [Google Scholar]
- 14.Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS. Model system for plant cell biology: GFP imaging in living onion epidermal cells. BioTechniques. 1999;26:1125–1132. doi: 10.2144/99266st04. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]