Abstract
Phytomers are developmental compartments that display stereotypical patterns dependent on whether they are initiated during the vegetative phase or the floral phases. Differences in appearance result from differential partitioning mechanisms responsible for allocation of cells to different components of the phytomer. The tasselsheath loci of maize control cell partitioning within the phytomer, indirectly influencing growth and development of its individual components. The tasselsheath4 (tsh4) gene accomplishes this through regulation of the ramosa2 (ra2) meristem determinacy gene, whereas tasselsheath1 (tsh1) appears to function differently.
Key words: meristem, SBP box, bract, maize, ramosa, tasselsheath, phytomer
Aerial plant development is organized into repeating units called phytomers, each consisting of three components, an axillary meristem, subtending leaf and associated internode.1 While vegetative phytomers and floral phytomers have similar organization, there are differences in the degree of elaboration of the different components. For example, within vegetative phytomers the leaf portion is often noticeable and highly elaborated. In floral phytomers, however, the leaf portion is highly reduced and obscured, and the axillary meristems are elaborated, eventually forming the reproductive organs (Fig. 1A). Clonal analysis of maize development has shown that sectors tend to begin and end within phytomer boundaries, indicating that the phytomer behaves as a developmental compartment.2 Thus, the cells of the phytomer share a common origin, and developmental mechanisms to partition these cells into the different components of the phytomer must exist.
Figure 1.
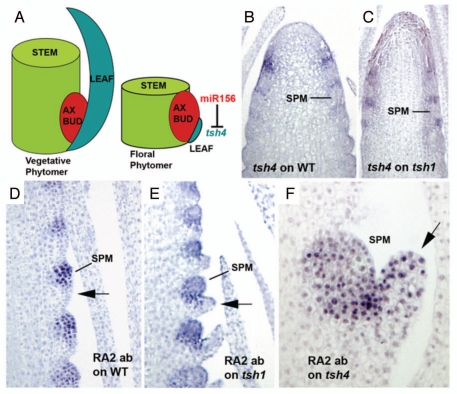
(A) Schematic of phytomer organization within the vegetative and floral phases. The two phases differ by the degree of elaboration of the three components. Within the floral phase of maize, the leaf component is repressed by the action of the tsh4 gene, which itself is repressed by meristem specific expression of miR156. (B) tsh4 in situ hybridization on wildtype tassels showing expression subtending SPMs. (C) tsh4 in situ hybridization on tsh1 tassels showing expression similar to wildtype. (D) Immunolocalization of RA2 in wildtype tassels. RA2 is found throughout the SPM, but not in the subtending repressed bract (arrow). (E) Immunolocalization of RA2 in tsh1 tassels. No ectopic RA2 is found in the de-repressed bract (arrow). (F) Immunolocalization of RA2 in tsh4 tassels. Ectopic RA2 is found in the de-repressed bract (arrow).
Recently, a new class of mutants in maize has been described that confirmed the existence of these developmental mechanisms within the phytomer. The tasselsheath (tsh) mutants cause de-repression of bract leaf development within the floral phytomers.3 This bract leaf growth occurs at the expense of the axillary meristems of the inflorescence, including the branch meristems (BM) that produce the side branches of the tassel and the spikelet pair meristems (SPM) that produce the spikelets. Both of these meristems are either reduced in number in the tasselsheath mutants, or have reduced determinacy and thus produce fewer lateral primordia. To date, two tasselsheath loci have been described, tsh1,4 and tsh4.5 Loss-of-function mutants of both genes display nearly identical phenotypes. tsh1 is a GATA class transcription factor that has conserved function and expression patterns in several grasses. tsh1 is expressed at the base of the SPM in areas where repressed bract leaves exist, indicating that the gene functions to repress bract leaf development during the floral phase.4 tsh4 is a microRNA regulated SBP box transcription factor that is also expressed in these areas (Fig. 1B), but has a wider expression pattern in the stem.5 Interestingly, both tsh1 and tsh4 mutants display numerous defects in axillary meristem development, and yet neither gene is expressed in meristems. Concerning tsh4, previous studies have shown that one explanation for this observation is that factors that repress it, including the miR156 microRNA, are expressed within meristems and function to turn it off. In support of this, double labeling experiments of miR156 RNA and TSH4 protein show a complementary expression pattern,5 where the microRNA is localized in the axillary meristem and TSH4 is localized to the suppressed bract leaf (Fig. 1A). tsh1/tsh4 double mutants appear indistinguishable from the single mutants (data not shown), making epistasis difficult to establish. tsh4 shows a normal pattern of expression within tsh1 inflorescences (Fig. 1C), indicating that tsh1 is not required for tsh4 expression.
In order to understand how tsh4 can affect axillary meristems without being expressed in them, the expression of lateral meristem markers was observed in tsh4 mutants. One such marker is the ra2 gene that functions to specify SPM determinacy. ra2 is a LOB domain transcription factor expressed within the SPM, but not in the bract subtending it6 (Fig. 1D). Expression of RA2 protein within tsh1 appeared normal (Fig. 1E). However, ectopic RA2 protein was observed within the de-repressed bracts of the tsh4 mutant (Fig. 1F). These results indicate that tsh4 plays a specific role in repressing ra2 expression within bracts that is distinct from tsh1. The ectopic RA2 protein within the tsh4 bracts reflect lost axillary meristem potential, indicating that de-repressed bract growth occurs at expense of axillary meristem growth, i.e., cells normally destined for the SPM are reallocated into the bract. This observation may explain how tsh4 can affect axillary meristem growth without actually being expressed within them. How tsh1 is able to accomplish the same function independent of ra2 is unclear.
Overall, these results show that a limited number of cells initially form the phytomer compartment, and that competition for these cells exists between the individual phytomer components. For example, the bract component competes with the axillary meristem component for cells. When the bract is de-repressed, its growth re-allocates cells normally destined to form the axillary meristem, as shown by the fact that bracts in tsh4 mutants ectopically express meristem markers such as ra2 (Fig. 1D). Since previous studies of tsh4 have shown that the antagonism between cells of the axillary bud and the subtending bract may be mediated by miR156, the possibility remains that meristem specific genes such as ra2 may regulate microRNA expression.
Acknowledgements
This work was supported by DOE grant DE-A102-08ER15962.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12220
References
- 1.Rutishauser R, Sattler R. Complementarity and heuristic value of contrasting models in structural botany. Bot Jahrb Syst Pflanzengesch Planzengeogr. 1985;107:415–455. [Google Scholar]
- 2.Johri MM, Coe EH. Clonal analysis of corn plant development. I. The development of the tassel and the ear shoot. Dev Biol. 1983;97:154–172. doi: 10.1016/0012-1606(83)90073-8. [DOI] [PubMed] [Google Scholar]
- 3.McSteen P, Hake S. barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development. 2001;128:2881–2891. doi: 10.1242/dev.128.15.2881. [DOI] [PubMed] [Google Scholar]
- 4.Whipple C, Hall D, DeBlasio S, Taguchi-Shiobara F, Schmidt R, Jackson D. A conserved mechanism of bract suppression in the grass family. Plant Cell. 2010 doi: 10.1105/tpc.109.073536. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuck G, Whipple C, Jackson D, Hake S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010;137:1243–1250. doi: 10.1242/dev.048348. [DOI] [PubMed] [Google Scholar]
- 6.Bortiri E, Chuck G, Vollbrecht E, Rocheford TF, Martienssen R, Hake S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell. 2006;18:574–585. doi: 10.1105/tpc.105.039032. [DOI] [PMC free article] [PubMed] [Google Scholar]