Abstract
Plants exposed to stress pass the memory of exposure to stress to the progeny. Previously, we showed that the phenomenon of transgenerational memory of stress is of epigenetic nature and depends on the function of Dicer-like (DCL) 2 and DCL3 proteins. Here, we discuss a possible role of DNA methylation and function of small RNAs in establishing and maintaining transgenerational responses to stress. Our new data report that memory of stress is passed to the progeny predominantly through the female rather than male gamete. Possible evolutionary advantages of this mechanism are also discussed.
Key words: transgenerational response to stress, Arabidopsis thaliana, maternal inheritance, methylation changes, homologous recombination frequency, genome instability, adaptive response, dcl2, dcl3
Plants are sedentary organisms and thus can not respond to rapidly changing growth conditions by escaping to new environments as animals usually do. Moreover, since seed dispersal is rather limited in the vast majority of plants, the progeny is very likely to grow under the same environmental growth conditions as its parents did. The memory of pre-existing growth conditions can be advantageous for plant survival. The environmental experience of parents can be recorded in the form of induced epigenetic modifications that occur in somatic cell lineages. The very late, almost at the end of plant development, separation of germline cells from somatic tissues enables incorporation of acquired epigenetic changes in the gametes. Indeed, previous reports suggested that the progeny of exposed plants might have an advantage while growing in the same environment as its parents.1–3 Despite a growing number of experimental evidences that support the existence of the phenomenon of memory of stress, the data on adaptive changes in the progeny of stressed plants are scarce.
Parental exposure to stress may not only lead to adaptive effects in progeny but also introduce a certain degree of changes in genome stability.4–9 Our early report showed that the progeny of tobacco plants infected with tobacco mosaic virus had an increased meiotic recombination frequency.8 A more recent report demonstrated that these progeny plants had a higher frequency of rearrangements at the loci carrying the homology to N-gene-like R-gene loci, allowing speculations about a possible role of these rearrangements in pathogen resistance evolution.9 Similarly, a study of Molinier et al. (2006) showed that the progeny of plants exposed to UVC or flagellin had an increased frequency of somatic homologous recombination events (HRF).4 The authors demonstrated that an increase in HRF triggered by a single exposure to UVC was maintained for five consecutive generations in the absence of stress. In contrast, our most recent reports demonstrated that maintaining an increase in HRF caused by ancestral exposure to heat, cold, flood, UVC or salt required exposure to stress in subsequent generations: if F1 plants were propagated for one more generation without stress, the effect diminished and HRF returned back to the level observed in the progeny of untreated plants.6,7 This scenario seems to be more probable from an evolutionary point of view. Within a given environmental niche, plants establish certain genetic and epigenetic traits needed to cope with the expected growth conditions. Drastic environmental changes or new unusual stresses may trigger a cascade of gene expression changes in attempt to survive and adapt to new conditions. Some of these potentially advantageous changes are most probably recorded in the form of DNA methylation and chromatin modifications and are passed to progeny as memory of stress exposure.
It can be further hypothesized that if these new environmental conditions are no longer present during the lifespan of future generations, the newly established methylation patterns and chromatin organization will return to the original epigenetic landscape that was the most adequate fit for this environmental niche. If the same new stresses occur in consecutive generations, the newly established epigenetic changes will be maintained and possibly stabilized after many generations of exposure.
Maternal Gametes Contribute more Significantly to Transgenerational Changes
Transmission of transgenerational changes could be either maternal/paternal or biparental in origin. Since the female gamete contributes the majority of cytoplasm to a zygote, it is more likely that changes occurred in the maternal epigenome and consequently in transcriptome and metabolome will be passed to progeny. As the majority of DNA methylation is erased shortly after fertilization and thus needs to be re-established, the pool of maternally inherited smRNA can facilitate the incorporation of epigenetic changes that were recently acquired by mother into the embryo epigenome. Indeed, recent studies by Mosher et al. (2009) have provided strong evidence that a large number of PolIV-dependent smRNAs are maternally expressed and are already accumulated in plant gametophyte before fertilization.10 Noteworthy, the presence of several classes of smRNA was also observed in male gametophyte and sperm cells,11,12 suggesting that paternal transmission of stress memory is also possible. While memory of the maternal environmental experience (including nutrients availability, light intensity, shade positioning, etc.,) can be of minor importance to self-pollinating species, it will be of advantage to the progeny of cross-pollinating species as seed dispersal is rather limited when compared to pollen.
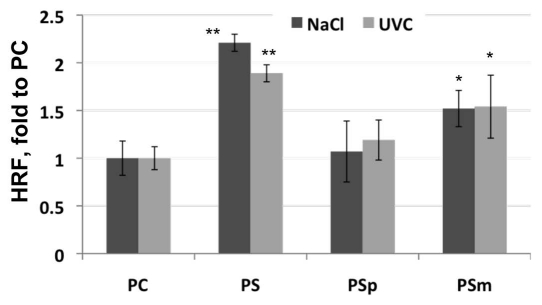
To analyze the parental contribution towards stress-induced transgenerational changes, we exposed Arabidopsis plants to two different stresses, UVC and salt. Next, control and stressed plants were selfed to obtain the progeny of control (PC) and stressed (PS) plants. In parallel, emasculated flowers of non-stressed control plants were pollinated manually with pollen obtained from stressed plants to generate the progeny of paternally exposed plants, PSp. Also, emasculated flowers of stressed plants were manually pollinated with pollen from non-stressed plants to obtain the progeny of maternally exposed plants, PSm. The analysis showed that somatic HRF was 2.21- and 1.89-fold higher in PS plants that were derived from plants exposed to UVC and salt stress as compared to PC plants. Interestingly, HRF in PSm plants derived from plants exposed to both UVC and salt stress was substantially higher as compared to that in the respective PSp plants. HRF in PSm plants obtained from plants exposed to UVC and salt was 1.52- and 1.54-fold higher than in PC plants. In contrast, PSp plants obtained from plants exposed to UVC and salt showed HRF that was only 1.13–1.19-fold higher as compared to PC plants. Thus, transmission of stress memory through the female gametes is more efficient than through the male gametes. In such case, the male gametes will contribute genetic diversity to the progeny, whereas the female gametes will contribute both genetic and epigenetic diversity. However, there are other factors that could also increase maternal contribution; they include larger maternal nuclear contributions to the endosperm and a maternal origin of sporophytic tissues surrounding the female gametophyte and seed. The epigenetic state of the parental genome may also play a role in progeny phenotype. A recent report has shown that differences in methylation in the female and male gametes have different effect on seeds of the progeny.13 Reciprocal crosses between met1, a mutant of DNA METHYLTRANSFERASE1 (MET1), a primary maintenance DNA methyltransferase and wild-type Arabidopsis plants caused parent-of-origin effects on F1 seed size.13 Large seeds were obtained if the maternal genome was hypomethylated, whereas small seeds were obtained if the paternal genome was hypomethylated. The same effects have been observed for ddm1, a mutant of DECREASE in DNA METHYLATION.
Deficiency of dcl2 and dcl3 in Transgenerational Response to Stress Suggests the Role of siRNA in the Process
Small RNA-mediated regulation of gene expression plays a key role in a large number of developmental, physiological and stress-related processes in plants.14–16 Furthermore, the existence of a stress-inducible pool of smRNAs supports their possible involvement in the establishment of stress-related epigenetic marks. As we discussed above, smRNAs may be among those molecular factors which, if present in the female gametophyte, would mediate predominant transmission of memory of maternal stress to progeny. If this hypothesis is correct, it would be safe to assume that plants deficient in smRNA biogenesis are also impaired in transgenerational response to stress. Since DCL2, DCL3 and DCL4 proteins show some functional redundancy and are required for biogenesis of several classes of smRNA, we studied a role of smRNAs in transgenerational response to stress using the respective single, double and triple mutants. In agreement with our hypothesis, the progeny of dcl2 and dcl3 mutant plants exposed to cold, heat, drought and UVC stress was partially impaired in an increase in somatic HRF. The role of epigenetic changes in transgenerational stress response was further supported by the absence of significant changes in DNA methylation in the progeny of dcl2 and dcl3 mutants grown under heat or UVC stress conditions as compared to the progeny of wild-type plants. This is not surprising, since the involvement of siRNAs which constitute the majority of smRNA pools generated by DCL proteins is well documented.17 Overall, our findings support the role of DCLs and siRNAs in transmission of stress memory to progeny.
Short interfering RNAs can be involved in the establishment of transgenerational changes at several different levels. They could mediate direct responses to stress, be responsible for generation of a stress signal and/or its propagation from somatic tissue to gametes and, which is more likely, they could guide the establishment of specific chromatin changes in gametes. Unfortunately, there are very few reports showing that siRNAs can possibly alter methylation patterns at specific genomic locations.18–20 At the same time, non-symmetrical genome methylation maintained by siRNA represents a very flexible epigenetic mark that may be quickly lost if a siRNA trigger is removed.17 This mechanism would make possible a fast introduction and removal of epigenetic marks in response to stress; it can also explain why transgenerational stress memory is nearly lost in the second generation after exposure to parental stress. In fact, a number of developmentally regulated genes are controlled via non-symmetrical cytosine methylation that enables their continuous reprogramming.18,21 On the contrary, CG methylation is known for robust transgenerational inheritance22 and is more resistant to reprogramming. Thus, it is less likely to be used as an epigenetic mark during transgenerational stress response observed in our study. Since siRNAs are believed to be primarily associated with an increase rather than a decrease in methylation, higher methylation levels observed in the progeny of stressed plants also support the involvement of the siRNA-directed epigenetic pathway in this process. Mutants impaired in CHROMOMETHYLASE3 (CMT3) and DOMAIN REARRANGED METHYLTRANSFERASES1 (DRM1), genes required for the maintenance of non-symmetrical cytosine methylation, accumulate a number of pleiotropic developmental effects. Therefore, it can be suggested that siRNA-induced changes in non-symmetrical methylation can indeed lead to changes in phenotype in the progeny.
It is possible that stress-generated siRNAs are transported to meristematic cells which later give rise to the germline. In fact, a recent report by Chitwood et al. (2009) supported the role of smRNAs as mobile signals during plant development.23 Predominant transmission of memory of maternal stress reported here may be explained by amplification of the received siRNA pool in the female rather than male gametes. Alternatively, it is also possible that lower contribution of the male siRNAs pool to the progeny's phenotype is determined by a very small volume of sperm cell cytoplasm compared to egg cell cytoplasm. Although our work has some indications that such mechanism of stress memory inheritance exists, it remains to be established whether there is a difference in smRNAs pools from gametes of stressed and non-stressed plants; and whether there is a link between these smRNAs populations and changes in DNA methylation and chromatin structure at genomic loci relevant to stress response. Even though we were not able to show direct correlation between the level of DNA methylation at specific loci and the frequency of recombination at those loci, we believe that such correlation should exist. Using deep sequencing technology, a more in-depth analysis should allow to study patterns of genome rearrangements and locus-specific DNA methylation. It would be important to establish a link between the presence of specific stress-induced siRNAs and changes in stability and DNA methylation at given genomic loci.
Figure 1.
To be exposed to UVC, plants were grown on soil and at 14 days post germination were irradiated with 7,000 ergs UVC. Control plants were grown under normal conditions. To be exposed to salt stress, plants were germinated and grown on MS medium supplemented with 25 mM NaCl. Control plants were grown on normal MS medium. At the age of three weeks plants were moved to soil. Control and stressed plants were selfed giving rise to PC and PS plants. Stressed plants were also used to cross-pollinate control plants in reciprocal manner giving rise to PSm and PSp plants. Five plants for each reciprocal cross were used. HRF was analyzed in the population of 50–100 plants at three weeks post germination. Asterisks show the difference in HRF as compared to PC plants (one—p < 0.05; two—p < 0.01).
Acknowledgements
The work was supported by grants from NSERC, HFSP and AARI.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12227
References
- 1.Johnsen O, Daehlen OG, Ostreng G, Skroppa T. Daylength and temperature during seed production interactively affect adaptive performance of Picea abies progenies. New Phytol. 2005;168:589–596. doi: 10.1111/j.1469-8137.2005.01538.x. [DOI] [PubMed] [Google Scholar]
- 2.Sultan SE. Phenotypic plasticity for plant development, function and life history. Trends Plant Sci. 2000;5:537–542. doi: 10.1016/s1360-1385(00)01797-0. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal AA, Laforsch C, Tollrian R. Transgenerational induction of defences in animals and plants. Nature. 1999;401:60–63. [Google Scholar]
- 4.Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- 5.Pecinka A. Transgenerational stress memory is not a general response in Arabidopsis. PLoS ONE. 2009;4:5202. doi: 10.1371/journal.pone.0005202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyko A, Blevins T, Yao Y, Golubov A, Bilichak A, Ilnytskyy Y, et al. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS One. 2010;5:9514. doi: 10.1371/journal.pone.0009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyko A, Golubov A, Bilichak A, Kovalchuk I. Chlorine ions but not sodium ions alter genome stability of Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1066–1078. doi: 10.1093/pcp/pcq048. [DOI] [PubMed] [Google Scholar]
- 8.Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, et al. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature. 2003;423:760–762. doi: 10.1038/nature01683. [DOI] [PubMed] [Google Scholar]
- 9.Boyko A, Kathiria P, Zemp FJ, Yao Y, Pogribny I, Kovalchuk I. Transgenerational changes in the genome stability and methylation in pathogen-infected plants: (virus-induced plant genome instability) Nucl Acids Res. 2007;35:1714–1725. doi: 10.1093/nar/gkm029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 11.Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, et al. MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics. 2009;10:643. doi: 10.1186/1471-2164-10-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL, et al. Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 2006;142:1160–1168. doi: 10.1104/pp.106.088849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 15.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyko A, Kovalchuk I. Epigenetic control of plant stress response. Environ Mol Mutagen. 2008;49:61–72. doi: 10.1002/em.20347. [DOI] [PubMed] [Google Scholar]
- 17.Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, et al. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769:358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE, et al. RNAi, DRD1 and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet. 2006;2:83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurihara Y, Matsui A, Kawashima M, Kaminuma E, Ishida J, Morosawa T, et al. Identification of the candidate genes regulated by RNA-directed DNA methylation in Arabidopsis. Biochem Biophys Res Commun. 2008;376:553–557. doi: 10.1016/j.bbrc.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. Pattern formation via small RNA mobility. Genes Dev. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]