Abstract
Waterlogging is a serious impediment to crop productivity worldwide which acts to reduce oxygen levels in the rhizosphere due to the low diffusion rate of molecular oxygen in water. Plants respond to low oxygen through rapid and specific changes at both the transcriptional and translational levels. Transcriptional changes to low-oxygen (hypoxia) stress have been studied in a number of plant species using whole genome microarrays. Using transcriptome data from root tissue from early time points (4–5 h) from cotton (Gossypium hirsutum), Arabidopsis and gray poplar (Populus x canescens), we have identified a core set of orthologous genes that responded to hypoxia in similar ways between species, and others that showed species specific responses. Responses to hypoxia were most similar between Arabidopsis and cotton, while the waterlogging tolerant poplar species exhibited some significant differences.
Key words: low-oxygen stress, hypoxia, abiotic stress, waterlogging, Arabidopsis thaliana, Gossypium hirsutum, cotton, Populus x canescens, gray poplar
In silico Comparisons of Hypoxia Stress between Cotton, Arabidopsis and Poplar
The growth and gene expression responses of cotton to waterlogging were recently assessed.1 Growth was found to slow and the speed of recovery was affected by the duration of the waterlogging stress. Gene expression profiles of key genes suggested that cotton may have differing strategies for responding to short and long term waterlogging events. To compare the cotton transcriptome waterlogging response with other dicotyledonous species, in silico comparisons of early hypoxia stress response (4–5 h) were made between cotton and Arabidopsis and poplar.
Affymetrix microarray data from three hypoxia experiments were used for comparison. The datasets analyzed came from cotton root tissue which had been submerged for 4 h1 (GSE16467), root tissue isolated from poplar which had their roots submerged for 5 h2 (GSE13109) and previously unpublished data from Arabidopsis roots subjected to a 0.1% O2 (balance N2) atmosphere for 5 h (GSE21504). Genes with significantly changed expression in response to hypoxia/waterlogging were determined using data processed in the same manner for each dataset.1 Those genes with a greater than two-fold change in expression and a false discovery rate adjusted p-value less than 0.05 were considered to be significant.
In order to compare results across species, gene homologues between species were identified. All cotton ESTs used to derive the microarray probes were compared to either the Arabidopsis TAIR9 Peptide database or the ESTs used to derive the poplar Affymetrix probes using BLASTX or TBLASTX.3 In each case the top three Arabidopsis or poplar hits to each cotton EST were identified to allow for the possibility that differing members of a gene family may fulfill the same roles in the different species.
Core Hypoxia Responses and Species Differences
A total of 1,012 cotton (593 upregulated, 419 downregulated), 2,386 Arabidopsis (1,175 upregulated, 1,211 downregulated) and 2,289 poplar (1,286 upregulated, 1,003 downregulated) genes were found to be differentially expressed in response to hypoxia (Suppl. Fig. 1). These genes represented 4.2%, 10.5% and 3.7% of the genes on the respective species microarray. Of the 24,132 cotton sequences represented on the Affymetrix array, 18,960, 19,232 and 18,018 had a good match (e-value < 1−10) to at least one Arabidopsis peptide sequence, translated poplar EST sequence or both, respectively.
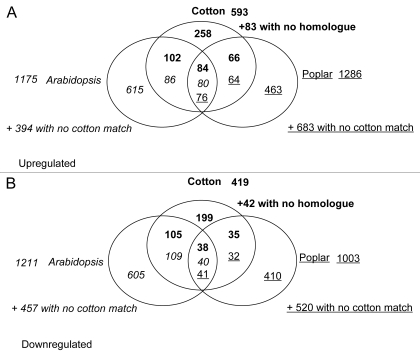
Comparing the responses to low-oxygen in all three species, more cotton waterlogging responsive genes had a similarly responsive Arabidopsis homologue than poplar homologue (Fig. 1). There were 84 upregulated cotton genes with homologues upregulated in both species and 38 downregulated cotton sequences with downregulated homologues in the other species (Fig. 1; Suppl. Table 1). These core low oxygen response genes included multiple members of the ethanolic fermentation genes ADH (2 genes) and PDC (5 genes) respectively. Also included were 11 cotton genes with putative roles in sugar metabolism and glycolysis, including sucrose synthase, hexokinase and phosphofructokinase homologues. A putative hemoglobin was also found in this core set. The largest category of genes (26/122) were those with putative transcription factor activity, including seven genes with similarity to ethylene response transcription factors. Also notable were 11 putative heat shock proteins, six putative proteases, five genes with potential roles in cell wall modification, three putative cytochrome P450 genes and two genes with similarity to lipases. The core set of genes identified here fits well with the set of core plant responses identified in a cross-kingdom comparison of hypoxic transcriptional responses, where fermentation, sucrose transport, reactive-oxygen species regulation, heat shock protein, and ethylene response factor associated genes were found to be differentially expressed in response to low-oxygen stress.4 Overall there was greater overlap in homologous gene responses between cotton and Arabidopsis (207 genes) than between cotton and poplar (101 genes; Fig. 1).
Figure 1.
Venn Diagram of overlap between hypoxia/waterlogging responsive homologues. (A) genes upregulated and (B) genes downregulated by hypoxia/waterlogging stress. Because cotton gene homologues in Arabidopsis and poplar were identified by collecting the top three blastx or tblastx matches to cotton EST sequences, it is possible for a given Cotton sequence to match multiple stress responsive homologues in another species or for multiple cotton sequences to match the same homologue in another species.
Given the difficulties in comparing inter-specific transcriptional responses on a gene-to-gene basis in plants where large gene families and duplicated genomes are common, functional category/pathway level was also used to assess differences in transcriptional responses to low oxygen between the different species. The PageMan software5 was used to compare those functional annotation bins in each species which had greater numbers of genes showing a transcriptional response larger than two-fold than could be expected by chance. Broadly, 71, 89 and 82 functional bins were found to be significantly active in Arabidopsis, cotton and poplar respectively (Table 1; Suppl. Fig. 2). 18 bins were commonly active in all three species and these included ‘fermentation’, ‘hormone metabolism.ethylene’, ‘stress abiotic’, ‘stress abiotic.heat’, ‘RNA.regulation of transcription.AP2/EREBP, APETALA2/Ethylene’ and ‘RNA.regulation of transcription.WRKY domain transcription factor’ which were upregulated and ‘cell wall’, ‘cell wall.degredation’, ‘secondary metabolism’, ‘secondary metabolism.phenylpropanoids’, ‘signalling.receptor kinases’, ‘signaling. receptor kinases.leucine rich repeat III’ and ‘transport’ which were downregulated. In both cotton and Arabidopsis roughly 2 times as many functional bins were downregulated as were upregulated (62:27 and 46:25 respectively; Table 1; Suppl. Fig. 2). In contrast the response in poplar skewed more towards transcriptional activation of functional bins (33:49; Table 1; Suppl. Fig. 2). Functional bins associated with cell wall synthesis and degradation were downregulated significantly in Arabidopsis and cotton as compared to poplar (8, 11 and 3, respectively).
Table 1.
Summary of pageman functional bins affected by hypoxic stress in cotton, Arabidopsis and poplar
| Functional bin | Number of sub-bins with upregulated genes over-represented | Number of sub-bins with downregulated genes over-represented | ||||
| Cotton | Arabidopsis | Poplar | Cotton | Arabidopsis | Poplar | |
| major CHO metabolism | 2 | 0 | 1 | 1 | 0 | 0 |
| minor CHO metabolism | 0 | 1 | 3 | 0 | 0 | 1 |
| glycolysis | 2 | 0 | 1 | 0 | 0 | 0 |
| fermentation | 2 | 1 | 2 | 0 | 0 | 0 |
| cell wall | 0 | 0 | 2 | 11 | 8 | 3 |
| lipid metabolism | 0 | 0 | 1 | 1 | 0 | 1 |
| amino acid metabolism | 5 | 5 | 3 | 7 | 0 | 3 |
| secondary metabolism | 0 | 0 | 0 | 10 | 6 | 5 |
| hormone metabolism | 1 | 4 | 3 | 4 | 3 | 0 |
| stress | 3 | 3 | 4 | 2 | 1 | 0 |
| nucleotide metabolism | 2 | 0 | 0 | 0 | 1 | 0 |
| misc | 1 | 1 | 0 | 5 | 6 | 5 |
| RNA | 4 | 4 | 12 | 2 | 1 | 0 |
| DNA | 1 | 2 | 1 | 0 | 2 | 0 |
| Protein | 3 | 3 | 9 | 7 | 3 | 3 |
| signalling | 0 | 0 | 1 | 4 | 6 | 6 |
| development | 0 | 0 | 2 | 0 | 0 | 0 |
| transport | 0 | 0 | 1 | 2 | 5 | 2 |
| not assigned | 0 | 0 | 3 | 3 | 4 | 3 |
The poplar response to waterlogging included greater upregulation of trehalose-6-phosphate phosphatase genes, which are involved in trehalose synthesis, than observed in Arabidopsis and cotton (Suppl. Fig. 2). Trehalose has been shown to confer tolerance to multiple stresses in rice.6 Increased trehalose-6-phosphate inhibited SNF1 related protein kinase 1 activity leading to downstream increases in glycolysis, TCA cycle, mitochondrial electron transport, amino acid synthesis, protein synthesis and nucleotide synthesis gene expression.7 Trehalose synthesis and degradation genes have previously been noted to be upregulated in response to hypoxia in Arabidopsis where it was proposed they play a role in controlling sugar flow to glycolysis.8 A number of functional bins associated with protein degradation were also upregulated only in poplar (Suppl. Fig. 2), suggesting that protein degradation may be higher in poplar than in cotton or Arabidopsis. It has been observed that protein concentrations decreased in roots of waterlogged poplar and oak which are tolerant and moderately tolerant of waterlogging respectively, but remained high in short-term waterlogging treatments in the waterlogging sensitive beech.9
Interestingly, while amino acid metabolism was affected in all three species assessed, there was almost no overlap between species in their particular response. Both cotton and poplar saw increases in transcripts for genes associated with homoserine synthesis and decreases in transcripts for genes involved in synthesis of aspartate family amino acids, specifically methionine. Cotton also underwent an increase in transcripts for genes involved in synthesis of cysteine, degradation of leucine and downregulation of genes involved in the synthesis of asparagine and lysine. Poplar also saw an increase in proline synthesis associated transcripts and a decrease in phenylalanine synthesis associated transcripts. Arabidopsis alone saw an increase in genes associated with alanine synthesis. Amino acid levels were studied in poplar following flooding and it was observed that the relationship between transcript levels for genes involved in amino acid metabolism and levels of their corresponding amino acids was not always clear and that transcriptional changes seemed to respond to changes in metabolite level.2
All three species show an upregulation of genes involved in ethylene production and signalling which is known to be important for hypoxia tolerance.10,11 However, there are also differences in hormone responses between the species. Cotton decreased transcription of genes induced by gibberellin and of genes involved in jasmonate production and response. The decrease in jasmonate production and responsiveness may be linked to an observed decrease in biotic stress associated genes in cotton, as jasmonate plays a role in mediating plant responses to pathogens.12 In contrast, Arabidopsis shows a bimodal response, with both a strong increase in some Auxin responsive genes and a strong decrease in others. Auxin mediates growth of adventitious roots in Arabidopsis,13 which are produced in response to hypoxia in Arabidopsis.14
Conclusion
Analysis of microarray data from three dicotyledonous plant species subjected to waterlogging or low-oxygen stresses has identified common responses, but also highlighted differences. In particular, at an early stress time-point, a waterlogging tolerant poplar variety showed more differences in response than Arabidopsis and cotton. Some of these differences include a delay in the downregulation of cell wall metabolism associated genes and increase in genes associated with trehalose and trehalose-6-phosphate metabolism. There are also differences between all three species in the areas of hormone metabolism, and strikingly, amino acid metabolism. The similarities and differences observed in waterlogging responses provide an experimental framework to understand the importance of pathways to hypoxia tolerance.
Acknowledgements
This work has been supported by CottTech-a research alliance between CSIRO, Cotton Seed Distributors and the Australian Cotton Research and Development Corporation.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12231
Supplementary Material
References
- 1.Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW. Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.) Plant Cell Physiol. 2010;51:21–37. doi: 10.1093/pcp/pcp163. [DOI] [PubMed] [Google Scholar]
- 2.Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, et al. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 2009;149:461–473. doi: 10.1104/pp.108.125989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, et al. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010;152:1484–1500. doi: 10.1104/pp.109.151845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, et al. PageMan: an interactive ontology tool to generate, display and annotate overview graphs for profiling experiments. BMC Bioinformatics. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, et al. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA. 2002;99:15898–15903. doi: 10.1073/pnas.252637799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, et al. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 2009;149:1860–1871. doi: 10.1104/pp.108.133934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu FL, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J. Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 2005;137:1115–1129. doi: 10.1104/pp.104.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreuzwieser J, Furniss S, Rennenberg H. Impact of waterlogging on the N-metabolism of flood tolerant and non-tolerant tree species. Plant Cell Environ. 2002;25:1039–1049. [Google Scholar]
- 10.Hattori Y, Nagai K, Furukawa S, Song X-J, Kawano R, Sakakibara H, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- 11.Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 12.Vijayan P, Shockey J, Levesque CA, Cook RJ, Browse J. A role for jasmonate in pathogen defense of Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7209–7214. doi: 10.1073/pnas.95.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell. 2005;17:1343–1359. doi: 10.1105/tpc.105.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis MH, Dennis ES, Peacock WJ. Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol. 1999;119:57–64. doi: 10.1104/pp.119.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.