Abstract
In Arabidopsis thaliana, heat shock factor binding protein (AtHSBP) is a negative regulator of the heat shock response (HSR), and defective AtHSBP leads to seed abortion. We found that the wild-type and AtHSBP-knockout plants did not differ in ovule phenotypes at flower position 3, which indicates that the seed abortion occurs after fertilization and during embryogenesis. The conserved residues of the hydrophobic heptad repeat (HR) domains in AtHSBP were mutated and examined for their subcellular localization and interacting ability with heat shock factors (AtHSFs). The HR domains at the C terminus of AtHSBP are important for retaining AtHSBP in the cytoplasm under normal growth conditions and for interacting with AtHSFs, which negatively affects the DNA-binding capacity and transactivation activity of AtHSFs during the HSR.
Key words: embryogenesis, HSF, HSBP, HSR, seed development, thermotolerance
Under heat stress/shock (HS), cells have the potential to resist thermal damage and elevate the levels of heat shock proteins (HSPs) functioning as molecular chaperons to prevent protein denaturation and aggregation, which is called HS response (HSR). HSP expression is mediated by HS transcriptional factors (HSFs). At the beginning of the HSR, inactive HSFs translocate from the cytoplasm to the nucleus and become activated, and then bind to the HS element (HSE) of HSP promoters to regulate their expression. Within the attenuation of the HSR, HSP70 and heat shock factor binding protein (HSBP) directly associate with and inactivate HSFs to shut down the HSR.1–4
Members of the HSBP family are small proteins (<10 kD) and are highly conserved among species.5 Human HSBP1 (HsHSBP1) has been the first identified HSBP; predominantly nuclear-localized HsHSBP1 functions as a negative regulator of the HSR.3 In maize (Zea mays), EMPTY PERICARP 2 (EMP2), encoding the first described HSBP-like protein, regulates embryogenesis and shoot development. The emp2 mutant kernels show greatly increased HSP expression.6,7 In addition to EMP2, maize has another HSBP paralog, ZmHSBP2. Both ZmHSBP paralogs interact non-redundantly with specific HSFs, so EMP2 and ZmHSBP2 may have distinct functions during plant development and the HSR.7,8 In Arabidopsis, AtHSBP functions as a negative regulator during the recovery from HS. Cytosolic-localized AtHSBP depends on HS to translocate to the nucleus and inhibits AtHSFs binding to HSEs to attenuate the HSR. Altered levels of AtHSBP lead to differential HSP expression, and the defective AtHSBP also results in an aborted seed phenotype, but how AtHSBP influences seed development remains unclear.9
The Defective AtHSBP May Not Affect Ovule Development but May Influence Embryogenesis After Fertilization to Result in Seed Abortion
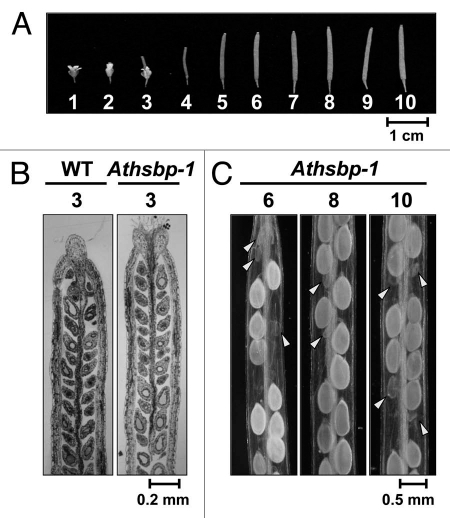
We examined the ovules in the ovary of different Arabidopsis flower positions that reflect the relative developmental stages on an inflorescence from the top to the bottom10,11 (Fig. 1A). Flower position 1 represents the opened youngest flower with visible petals between sepals. Flower positions 1 and 2 are self-pollinated flowers, and pollen tubes grow to fertilize the ovules. At and after flower position 3, the ovules enter embryogenesis and seed development. Semi-thin sections of the wild-type and AtHSBP-knockout (Athsbp-1) plants revealed no difference in ovule phenotypes at flower position 3 (Fig. 1B), whereas seed abortion phenotype was observed at and after flower position 6 (Fig. 1C). Thus, AtHSBP may not affect ovule development but may regulate embryogenesis and seed development after fertilization.
Figure 1.
Seed abortion phenotype in the AtHSBP-knockout line (Athsbp-1). (A) Flower positions from 1 to 10 as indicated. (B) Semi-thin sections of the ovary of the wild type (WT) and Athsbp-1 at flower position 3 were stained by toluidine blue O. (C) Seed abortion in green siliques at flower positions 6, 8 and 10. Arrowheads indicate aborted seeds.
The HSR is known to affect various developmental stages among different species. The emp2 mutant kernels in maize lead to unattenuated HSR and are also aborted at the coleoptile stage/stage 1, followed by necrosis and reabsorption of kernel contents.6 Overexpression of HSP70 in Drosophila cells has been found to reduce cell division rate and growth.12 We found that the defective AtHSBP upregulated HSP expression9 and affected embryogenesis to cause a seed abortion phenotype, which indicates that the mutant seeds of Athsbp-1 may exhibit an unattenuated HSR to cause aborted seeds. However, which embryonic stage is affected by AtHSBP needs to be confirmed.
Characterization of the AtHSBP Motif for Subcellular Localization and Interaction with AtHSFs
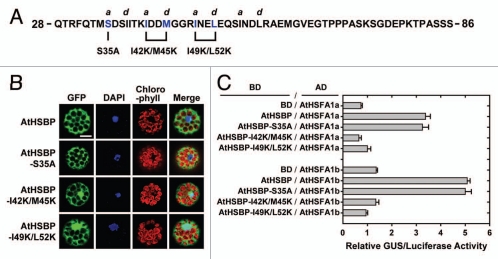
The coiled-coil region of HSBP is important for associating with the oligomerization domain of HSFs.3,8 The residues 41 to 65 of AtHSBP have been predicted to have 60% to 99% possibility to form a coiledcoil structure.9 Therefore, we mutated the conserved hydrophobic heptad repeat (HR) residues in this region to strong basic and hydrophilic residues, AtHSBP-I42K/M45K and AtHSBP-I49K/L52K, and the residue of Ser35 was mutated to Ala (AtHSBP-S35A), as shown in Figure 2A. These variant mutants were examined for their subcellular localization and interaction with AtHSFs.
Figure 2.
AtHSBP mutagenesis, cellular localization and interaction with AtHSFs. (A) The partial protein sequence of AtHSBP, residues 28 to 86. The hallmark of coiled-coil structure is a heptad repeat of 7 amino acid residues (“abcdefg”), with a predominance of hydrophobic residues at a and d positions, and residues at e and g positions are frequently charged, as predicted by COILS (http://www.ch.embnet.org/software/COILS_form.html) in a 14-residue window. The AtHSBP-S35A, AtHSBP-I42K/M45K and AtHSBP-I49K/L52K are shown below the sequence. (B) AtHSBP and the mutations were transiently expressed in Arabidopsis mesophyll protoplasts under normal growth conditions. Green fluorescent protein (GFP) signals were observed on confocal microscopy. Blue shows the nucleus stained with 4,6-diamino-phenylindole (DAPI), and red shows the chlorophyll with auto-fluorescence. Similar results were obtained from 3 independent replicates, and representative images are shown. Bar = 20 µm. (C) Quantification of protein-protein interaction by P2H assay. AtHSBP and the mutations were fused with GAL4 DNA-binding domain (BD) or -activation domain (AD), respectively, and used for protoplast transfections, as indicated. Transfections with BD and AtHSFs-AD constructs were used as references. Interactions between AtHSBP and AtHSFs were used as positive controls. The amount of relative GUS activity was normalized by luciferase luminescence. Data are means ± SD (n = 3).
AtHSBP and the mutants were fused to N terminus of green fluorescent protein (GFP), respectively, driven by a 35S promoter and analyzed in Arabidopsis mesophyll protoplasts to assess subcellular localization under normal growth conditions. AtHSBP-GFP and AtHSBP-S35A-GFP were dominantly expressed in the cytoplasm, whereas AtHSBP-I42K/M45K-GFP and AtHSBP-I49K/L52K-GFP were expressed both in the cytoplasm and nucleus (Fig. 2B). We also examined whether these mutants affect the interaction ability with the well-studied and HS-related AtHSFs, AtHSFA1a and AtHSFA1b, by P2H assays13 (Fig. 2C). The results confirmed that both AtHSBP-I42K/M45K and AtHSBP-I49K/L52K significantly reduced the interaction capability with AtHSFs, as compared with AtHSBP and AtHSBP-S35A.
Conclusions and Perspectives
During the recovery from HS, AtHSBP, without a nuclear localization signal, translocates to the nucleus and functions to negatively mediate the DNA-binding capacity and transactivation activity of AtHSFs.9 The conserved residue Ser31 of HsHSBP1 has been suggested to play an important role in its structure and function.14 Regardless of HS treatment, we found that the AtHSBP-S35A mutation may alter its structure for expression only in the cytoplasm, but does not affect the interacting ability with AtHSFs.9 The AtHSBP-I42K/M45K and AtHSBP-I49K/L52K mutations on the HR domain were detected both in the cytoplasm and nucleus, similar to that of the overexpressed GFP control (data not shown), and significantly lost the interacting activity with AtHSFs. We propose that (1) AtHSBP may interact with the specific factor(s) through the coiled-coil structure for retention in the cytoplasm under normal growth conditions, and the mutations of the AtHSBP-HR domain may lose the interacting ability and lead the overexpressed proteins to diffuse to the nucleus; and (2) the residue Ser35 can confer a specific conformation/structure of AtHSBP, which may be essential to associate with HS-related factor(s) for nuclear localization during the recovery from HS. However, the potential cytoplasmic interacting factor(s) mediating AtHSBP translocation in response to the HSR remain unclear, and additional studies are necessary to address this issue.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12404
References
- 1.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 2.Pirkkala L, Nykänen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 3.Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 1998;12:1962–1974. doi: 10.1101/gad.12.13.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 5.Tai LJ, McFall SM, Huang K, Demeler B, Fox SG, Brubaker K, et al. Structure-function analysis of the heat shock factor-binding protein reveals a protein composed solely of a highly conserved and dynamic coiled-coil trimerization domain. J Biol Chem. 2002;277:735–745. doi: 10.1074/jbc.M108604200. [DOI] [PubMed] [Google Scholar]
- 6.Fu S, Meeley R, Scanlon MJ. Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell. 2002;14:3119–3132. doi: 10.1105/tpc.006726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu S, Scanlon MJ. Clonal mosaic analysis of EMPTY PERICARP2 reveals nonredundant functions of the duplicated HEAT SHOCK FACTOR BINDING PROTEINs during maize shoot development. Genetics. 2004;167:1381–1394. doi: 10.1534/genetics.104.026575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu S, Rogowsky P, Nover L, Scanlon MJ. The maize heat shock factor-binding protein paralogs EMP2 and HSBP2 interact non-redundantly with specific heat shock factors. Planta. 2006;224:42–52. doi: 10.1007/s00425-005-0191-y. [DOI] [PubMed] [Google Scholar]
- 9.Hsu SF, Lai HC, Jinn TL. Cytosolic-localized heat shock factor binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis. Plant physiology. 2010 doi: 10.1104/pp.109.151225. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleecker AB, Patterson SE. Last exit: senescence, abscission and meristem arrest in Arabidopsis. Plant Cell. 1997;9:1169–1179. doi: 10.1105/tpc.9.7.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, et al. Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:15629–15634. doi: 10.1073/pnas.0805539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feder JH, Rossi JM, Solomon J, Solomon N, Lindquist S. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992;6:1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- 13.Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, et al. Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 2006;46:890–900. doi: 10.1111/j.1365-313X.2006.02731.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Xu L, Liu Y, Tong X, Zhu G, Zhang XC, et al. Crystal structure of the hexamer of human heat shock factor binding protein 1. Proteins. 2009;75:1–11. doi: 10.1002/prot.22216. [DOI] [PubMed] [Google Scholar]