Abstract
Through map-based cloning we determined TRICHOME BIREFRINGENCE (TBR) belongs to a plant-specific, yet anonymous gene family with 46 members in Arabidopsis thaliana. These genes all encode the domain of unknown function 231 (DUF231). TBR and its homolog TRICHOME BIREFRINGENCE-LIKE3 (TBL3) are transcriptionally coordinated with CELLULOSE SYNTHASE (CESA) genes, and loss of TBR or TBL3 results in decreased levels of crystalline secondary wall cellulose in trichomes and stems, respectively. Loss of TBR or TBL3 further results in increased pectin methylesterase (PME) activity and reduced pectin esterification in etiolated Arabidopsis hypocotyls. Together, the results suggest that DUF231 proteins might function in the maintenance of pectin- and probably homogalacturonan esterification, and that this is a requirement for normal secondary wall cellulose synthesis, at least in some tissues and organs. Here we expand the discussion about the role of TBL/DUF231 proteins in cell wall biology based on sequence and structure analyses. Our analysis revealed structural similarities of TBR with a rhamnogalacturonan acetylesterase (RGAE) of Aspergillus aculeatus and the protein LUSTRIN A-LIKE (Oryza sativa). The implications of these findings in regard to TBL functions are discussed.
Key words: cellulose, cell wall, pectin, esterase, lustrin A, DUF231, TBL, Arabidopsis
In our original work we showed that, besides the DUF231 domain, TBR/TBL proteins also contain a second conserved, plant-specific domain, which has not been described previously, and which we named TBL domain.2 An interesting feature of this domain is the presence of a conserved glycine-aspartate-serine (GDS) signature, which in the same amino acid context (i.e., GDSL) has previously been found to be a conserved motif in some esterases/lipases.1,10 Moreover the DUF231 domain contains a highly conserved amino acid stretch (DCXHWCLPGXXDXWN) towards the C-terminus of the proteins.2 A DXXH motif (as found at the beginning of this stretch) together with the Ser residue in the distant GDS motif in turn have been described to form the catalytic triad in the 2.5 Å crystal structure of a rhamnogalacturonan acetylesterase (RGAE) from the plant pathogenic fungus Aspergillus aculeatus.4
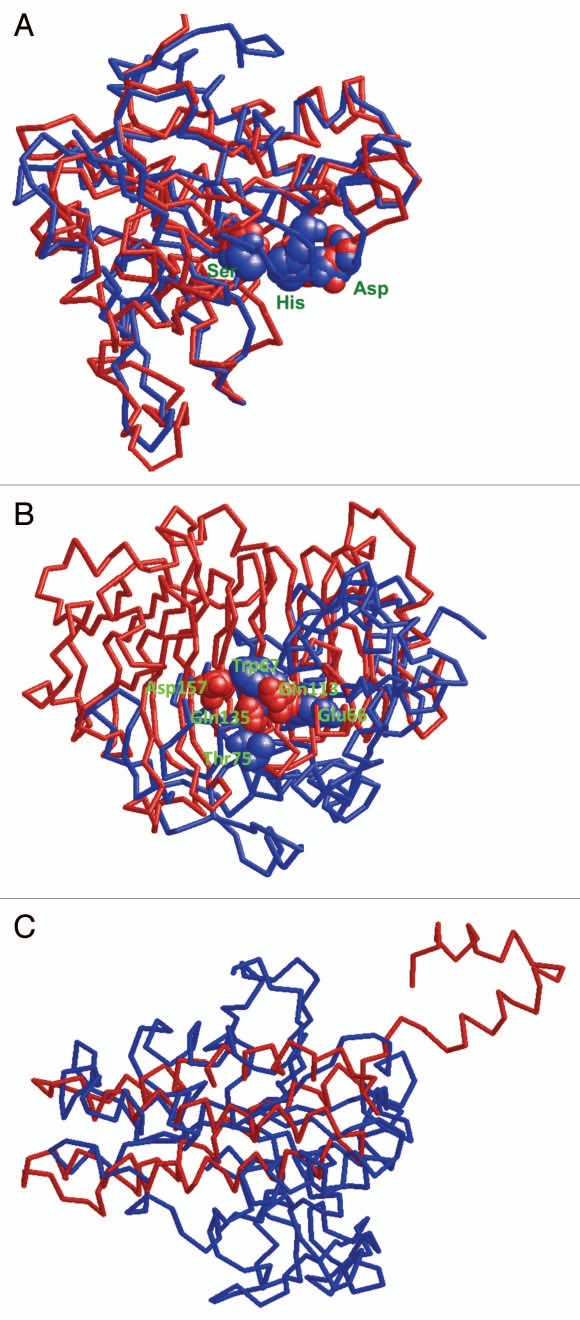
Following this observation, we produced an amino acid alignment of the fungal RGAE (Pfam database entry 1pp4) and the C-terminal half of TBR, with the N-terminus of RGAE corresponding to the hydrophobic amino acids preceding the GDS motif in TBR.2 223 amino acids of the 233 in the chain A of RGAE are involved in this alignment, which is characterized by 74 (23.1%) identical residues and another 136 (42.4%) similar residues. The predicted secondary structure of the corresponding fragment of TBR and the observed one of RGAE exhibit a similar succession of structural elements with the GDS motif being located at the end of a β-sheet and the DXXH motif residing in a loop region between two α-helices (not shown). Based on the alignment we then computed a spatial model for the TBR fragment using the RGAE structure as template (Fig. 1A).13 Remarkably, the predicted overall spatial structure of the TBR fragment and the positions of the amino acids (Ser, Asp, His) corresponding to the active site residues in RGAE show a near perfect match with the RGAE template (Fig. 1A).
Figure 1.
Modeled spatial structure of the TBR fragment. (A) Superposition of the backbones of the TBR fragment model (amino acids 325–604; red) with the RGAE structure (Aspergillus aculeatus; PDB: 1pp4, blue). Likely active site residues in TBR (Ser, His, Asp), which are conserved in the alignment with RGAE, are shown as space filling balls. (B) Superposition of the backbones of the TBR fragment model (blue) with the X-ray PME structure (Daucus carota; PDB: 1GQ8, red). The active site residues in PME (Gln113, Gln135, Asp136 not visible, Asp157) are shown as space filling balls. The aligned residues from TBR are Glu66, Trp67 and Thr75 (Rmsd = 7.7 Å, Z-Score = 1.6). (C) Superposition of the backbones of the TBR fragment model (blue) with the X-ray structure of PME-I (Arabidopsis thaliana; PDB: 1X8Z, red; Rmsd = 4.4 Å Z-Score = 3.5). Modeling was done with the program Jackal.13 The superposition was computed with CE.8 The figures were generated with RasMol.6
Despite this congruence at least two observations make it unlikely that TBL/DUF231 proteins have the same enzymatic function as RGAE or a general esterase activity. First, the crystallized RGAE from Aspergillus aculeatus belongs to the SGNH-hydrolases, a subclass of the GDSL enzymes.10 Besides the GDSL and DXXH motifs, SGNH hydrolases contain two additional interjacent conserved blocks with an invariable glycine (G) and asparagine (N).1 These two residues are close to the catalytic center in the crystal structure of RGAE4 and supposedly serve as proton donors,1 while they are absent in TBR/TBL proteins. Second, our experiments convincingly showed a reduced degree of pectin esterification in tbr and tbl3 mutant hypocotyls and significantly elevated PME activity,2 suggesting that TBR and TBL3 are required to maintain esterification of pectins. In compliance with this, we were also unable to find experimental evidence that supported an esterase function of TBR or TBL3 (data not shown).
Besides the spatial structural analogy between RGAE and the partial TBR, we could not find any striking sequence or structure similarities between TBR and known plant esterases such as the PME of Daucus carota (Fig. 1B). TBR neither features the plant PME-specific3 active center with its amino acid arrangement Asp136, Asp157, Gln113, Gln135, Arg225 (Daucus carota),3,5 nor the pectin-binding domain comprising several aromatic amino acids (Phe84, Tyr139, Phe160, Tyr222, Trp227, Phe250 and Trp252; Daucus carota).3 However, binding to pectin or cell wall polymers in more general might be indicated by the presence of several invariant cysteine and aromatic amino acid residues in the TBL and DUF231 domains;2 arrangements of cysteine and aromatic residues in a similar order are major determinants of cellulose or xylan binding modules CBM1 and CBM2 (cf. www.cazy.org). Also no structural similarities were found between the C-terminal half of TBR and PME inhibitors (PME-I), e.g., AtPME-I 1,11 (Fig. 1C).
TBR is annotated as being similar to Lustrin A-like (Oryza sativa; 52% identical residues and 68 positive residues). Lustrin A, like TBLs, contains the “GDSL” motif, situated in one of its ten cysteine-rich repeats, and has several other domains that were proposed to be important for elasticity between the aragonite layers in molluscs of the species Haliotis rufescens.12 Hence it has been proposed to be the adhesive component of the intercrystalline organic matrix lying between these layers of aragonite tablets.7,12 By analogy, this may suggest that TBR and TBLs are required to connect different layers of the plant cell wall.
The data in our original manuscript as well as the points discussed in this addendum make it unlikely that TBL/DUF231 proteins belong to a novel class of catalytically active pectin esterases. Our data rather promote the idea that TBL/DUF231 proteins represent either (1) catalytically inactive but pectin-binding proteins that, possibly by their sheer presence, curtail the activity of pectin esterifying enzymes or pectin esterases, functionally similar to polygalacturonase-inhibiting proteins,9 or (2) ‘bridging proteins’ that bind pectin and other cell wall polysaccharides, and hence crosslink different cell wall networks. The precise biochemical function of TBL proteins and biological functions for most family members remain to be determined.
Acknowledgement
Volker Bischoff is financed by a DFG grant BI 1417/1-1.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12414
References
- 1.Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. GDSL family of serine esterases/lipases. Prog Lipid Res. 2004;43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff V, Nita S, Neumetzler L, Schindelasch D, Urbain A, Eshed R, et al. TRICHOME BIREFRINGENCE and its homolog At5g01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis thaliana. Plant Physiol. 2010;153:590–602. doi: 10.1104/pp.110.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson A, El-Ahmad M, Friemann R, Jörnvall H, Markovic O, Eklund H. Crystal structure of plant methylesterase. FEBS Lett. 2002;514:243–249. doi: 10.1016/s0014-5793(02)02372-4. [DOI] [PubMed] [Google Scholar]
- 4.Mølgaard A, Larsen S. Crystal packing in two pH-dependent crystal forms of rhamnogalacturonan acetylesterase. Acta Crystallogr D Biol Crystallogr. 2004;60:472–478. doi: 10.1107/S0907444903029767. [DOI] [PubMed] [Google Scholar]
- 5.Pelloux J, Rustérucci C, Mellerowicz EJ. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007;12:267–277. doi: 10.1016/j.tplants.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Sayle R, Bissell A. RasMol: A Program for Fast Realistic Rendering of Molecular Structures with Shadows; Proc 10th Eurographics UK '92 Conference, University of Edinburgh; Scotland. 1992. [Google Scholar]
- 7.Shen XY, Belcher AM, Hansma PK, Stucky GD, Morse DE. Molecular cloning and characterization of lustrin A, a matrix protein from shell and pearl nacre of Haliotis rufescens. J Biol Chem. 1997;272:32472–32481. doi: 10.1074/jbc.272.51.32472. [DOI] [PubMed] [Google Scholar]
- 8.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Engin. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 9.Spadoni S, Zabotina O, Di Matteo A, Mikkelsen JD, Cervone F, De Lorenzo G, et al. Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of arginine and lysine. Plant Physiol. 2006;141:557–564. doi: 10.1104/pp.106.076950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Upton C, Buckley A new family of lipolytic enzymes. Trends in Biochem Sci. 1995;20:178–179. doi: 10.1016/s0968-0004(00)89002-7. [DOI] [PubMed] [Google Scholar]
- 11.Wolf S, Grsic-Rausch S, Rausch T, Greiner S. Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett. 2003;18:551–555. doi: 10.1016/s0014-5793(03)01344-9. [DOI] [PubMed] [Google Scholar]
- 12.Wustman BA, Weaver JC, Morse DE, Evans JS. Structure-function studies of the lustrin A polyelectrolyte domains, RKSY and D4. Connect Tiss Res. 2003;44:10–15. [PubMed] [Google Scholar]
- 13.Xiang Z, Honig B. Extending the accuracy limit of side-chain prediction. J Mol Biol. 2001;311:421–430. doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]