Abstract
Combination of molecular phylogenetic analyses of Chrysomelina beetles and chemical data of their defensive secretions indicate that two lineages independently developed, from an ancestral autogenous metabolism, an energetically efficient strategy that made the insect tightly dependent on the chemistry of the host plant. However, a lineage (the interrupta group) escaped this subordination through the development of a yet more derived mixed metabolism potentially compatible with a large number of new host-plant associations. Hence, these analyses on leaf beetles document a mechanism that can explain why high levels of specialization do not necessarily lead to “evolutionary dead ends.”
The vast majority of phytophagous insects are highly specialized in their feeding habits, such that they usually feed on a restricted set of host-plant species. This pattern of narrow association might be highly conservative in the course of evolution (1–3). Indeed, since the seminal paper of Ehrlich and Raven (4), many authors have supported the view that secondary plant substances strongly constrain phytophagous insects to shift among host plants that are chemically similar. Even radical shifts between two very distantly related plant families might be achievable only through the existence of a “phytochemical bridge” (5). Recent molecular phylogenetic analyses confirmed that secondary chemistry could explain host shifts better than plant phylogeny and plant geographic distribution (6, 7). In exceptional cases (8), the association between the insect and its host plant has even been conservative enough to yield highly correlated insect and host-plant phylogenies. Nevertheless, many host shifts in other phytophagous insects are not explained by phytochemical similarities (9). Clearly, there is a consensus emerging that, not only plant chemistry but also a suite of other parameters, influence host affiliation. Among them, biogeographical, genetic, and ecological constraints (10–13) are probably highly relevant.
Whatever the primary causes of specialization, a possible important corollary is that it could lead to an evolutionary dead end, i.e., specialization would cause a decrease in the ability of populations to answer new environmental conditions (e.g., requiring host shifts), hence, it would lead to a higher likelihood of extinction (14–20). This issue on the relative likelihood of survival for specialist and generalist species has been debated for decades (see ref. 10 for a review) and very few studies have specifically tested the question experimentally.
Chrysomelina leaf beetles constitute an excellent model to evaluate the ability of specialized insect herbivores to shift among hosts and to investigate the parameters allowing/preventing these changes. Indeed, most of the species belonging to this subtribe are highly specialized in their feeding habits. As the whole life cycle occurs on the same plant, a large number of parameters (feeding, reproduction, oviposition, larval development, chemical defense, etc.) are all influenced by a single host plant. When disturbed, chrysomeline larvae release repulsive volatile compounds secreted by dorsal glands. Qualitative analyses of Chrysomelina defensive secretions indicate three levels of specialization: full, partial, or lack of dependence of the insect chemical defense on the host-plant chemistry. Indeed, although many of these beetles de novo synthesize repulsive iridoid monoterpenes (21), a significant number of species exhibit defensive products whose synthesis is partly or completely dependent on the secondary compounds from their host. In the latter two cases, the insect defensive deterrents are, respectively, derived from sequestered plant toxins [i.e., salicylaldehyde is derived from Salicaceae phenolglycosides (22)], or produced by means of a mixed insect–plant biogenetic route [i.e., involving esterification of de novo-synthesized butyric acids by alcohols retrieved from the plant (23)]. Autogenous defense is encountered in species that specialize on any of the host-plant families reported for chrysomelines. On the other hand, toxins fully or partially derived from the host exclusively occur in larvae feeding on Salicacae or Betulaceae. Chrysomelina leaf beetles provide a pertinent model to test the order in which these three biogenetic routes evolved and influenced host affiliation.
Here, we report on a molecular phylogeny of Chrysomelina on which we mapped evolutionary changes in host affiliation and larval chemical defense strategies. The sampling of species was chosen to maximize the chemical diversity of defensive strategies to investigate whether higher host-dependence would lead herbivorous insects to evolutionary dead ends by reducing the possibility of further shifts to new host plants.
Materials and Methods
Data Collection.
Genomic DNA of species listed in Table 1 was extracted from ethanol-preserved single adult individuals by using standard protocols. We based our phylogenetic analyses on nucleotide sequences from the 12S mt ribosomal RNA (12S), the 16S mt ribosomal RNA (16S), the mt cytochrome oxidase I (COI), and the mt cytochrome oxidase II (COII) genes. Fragments from the 12S and 16S were sequenced from 35 Eurasian and American species [representing most (7/9) of the holarctic genera exhibiting defensive glands and belonging to the chrysomelina subtribe] listed in Figs. 1 and 2, whereas the COI and COII fragments were sequenced only for the species from the interrupta group, within which the ribosomal RNA divergences are extremely low (in analyses of the combined data set, COI and COII characters were coded as missing for the other taxa). Gonioctena variabilis, Chrysolina americana, and Oreina elongata were included as unambiguous outgroup taxa. The PCR-amplified mt DNA fragments were (i) a 502- to 553-bp segment of 12S (primers, SR-N-14759 and SR-J-14233, from ref. 24; 25010012Fc, 5′-ATTAGTATAAGATATGTTCTCG-3′; and 25010012Rc, 5′-CGATGTGTACATATTTTAGAGC-3′), (ii) a 423- to 500-bp segment of 16S (LR-J-12883 and LR-N-13398, from ref. 25; 25010016Fc, 5′-CTGCCCAATGATAATTGAATGG-3′; and 07010016SR1, 5′-CGCAATCTTTTCTTTCGATTTG-3′), (iii) a 620- to 649-bp segment of COI (C1-J-1751) and reverse complement of C-J-2441, both from ref. 24, and (iv) a 578- to 620-bp segment of COII (modTL2-J-3037, 5′-ATGGCAGATTAGTGCAWTRG-3′; and modC2-N-3661, 5′-CCACAAATTTCWGAACATTGACCA-3′, both modified from ref. 24). After a polymerase (AmpliTaqGold; Perkin–Elmer) activation step of 10 min at 94°C, thermocycling consisted of 35 cycles of 25 s denaturation, 60 s annealing (2 mM MgCl2), and 60 s extension, at 94°C, 51/52/53°C (for COI/COII/12S and 16S), and 72°C, respectively. This was followed by a final extension step of 7 min. PCR products were purified (QIAquick; Qiagen, Chatsworth, CA) and sequenced on both strands (dRhodamine Cycle Sequencing, electrophoresis on ABI 377; Applied Biosystems). Sequencing of complementary strands was performed on independent PCR products. Sequences were aligned with CLUSTAL W (26). Alignments of the protein-coding genes (COI and COII) were trivial and MACCLADE (v3.05) (27) was used to determine their ORFs. As alignment of 12S or 16S is notoriously difficult, even for moderately divergent sequences, we used the program SOAP (28) to produce one alignment for each of 25 different sets of alignment parameters (weighted matrix, gap penalties from 11 to 19 by steps of 2, and extension penalties from 3 to 11 by steps of 2). Positions at which alignments differed were excluded (28, 29). The single alignment produced with the default parameters of CLUSTALW was also analyzed to estimate whether it yielded results identical to those obtained after excluding unstable aligned positions. All sequences reported in this paper were deposited at GenBank under accession numbers AY027591–AY027628 and AY027695–AY027760.
Table 1.
Locality data of studied Chrysomelina specimens
| Group | Species | Locality data | |
|---|---|---|---|
| Ingroup | 1 | Chrysomela aenicollis | Colorado |
| 2 | C. collaris | Switzerland | |
| 3 | C. confluens | Utah | |
| 4 | C. cuprea | Switzerland | |
| 5 | C. falsa | Alaska | |
| 6 | C. interrupta | Maryland | |
| 7 | C. knabi | Utah | |
| 8 | C. lapponica-Cze | Czech Republik | |
| 9 | C. lapponica-Finl | Finland | |
| 10 | C. lapponica-MC | Massif Central, France | |
| 11 | C. lapponica-Que | Queyras, France | |
| 12 | C. laurentia | Pontiac, Canada | |
| 13 | C. mainensis interna | Alaska | |
| 14 | C. mainensis littorea | California | |
| 15 | C. mainensis mainensis | Pontiac, Canada | |
| 16 | C. populi | Belgium | |
| 17 | C. salicivorax | P. R. China | |
| 18 | C. schaefferi | California | |
| 19 | C. scripta | Ohio | |
| 20 | C. sp | Alaska | |
| 21 | C. tremulae | France | |
| 22 | C. vigintipunctata | France | |
| 23 | C. walshi | Pontiac, Canada | |
| 24 | Gastrophysa viridula | Belgium | |
| 25 | G. cyanea | Utah | |
| 26 | Linaeidea aenea | Belgium | |
| 27 | Phaedon brassicae | Germany | |
| 28 | Phratora vitellinae | Belgium | |
| 29 | P. tibialis | Belgium | |
| 30 | P. laticollis | Belgium | |
| 31 | Plagiodera versicolora | Belgium | |
| 32 | P. viridipennis | Brazil | |
| Outgroup | 33 | Chrysolina americana | France |
| 34 | Gonioctena variabilis | Spain | |
| 35 | Oreina elongata | France | |
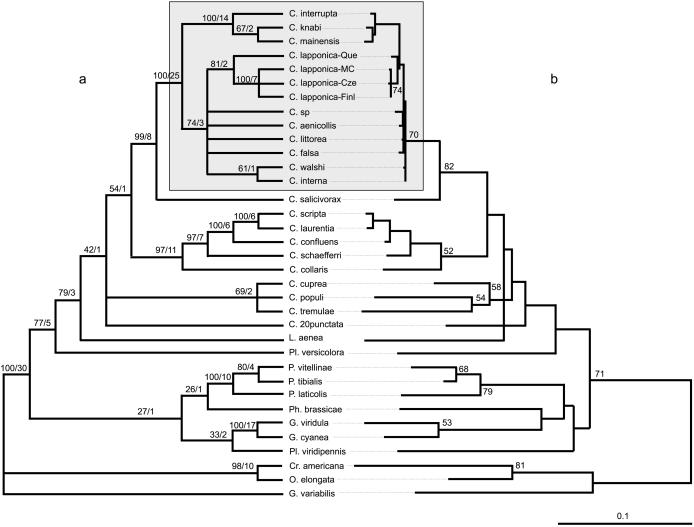
Figure 1.
(a) Strict consensus among the 120 MP trees (TL = 1532) with bootstrap values/decay indices indicated above the branches. (b) ML phylogram (−ln L = 10381.998, Ti/Tv = 1.734, Pinv = 0.421, γ shape parameter =0.536) with quartet puzzling support values >50% indicated above the branches. Scale is in percent expected substitution per position.
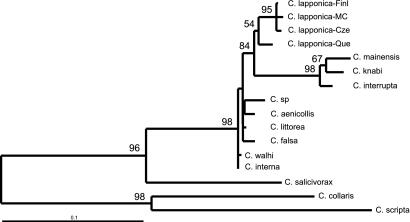
Figure 2.
ML phylogram of all species from the interrupta clade with a chosen subset of outgroup taxa. Numbers above the branches are bootstrap proportions >50% (400 replicates). Scale is in percent-expected substitution per position.
Phylogenetic Analyses.
We checked for possible saturation of nucleotide substitution types by plotting the number of transversions (Tv) vs. the number of transitions (Ti) as well as Ti and Tv vs. Tamura–Nei pairwise distances. Saturation plots were also examined separately for first, second, and third positions of protein-coding genes. All plots were used as a guide to develop weighted parsimony strategies. A 5% χ2 test comparing the nucleotide composition of each sequence to the frequency distribution assumed in the maximum likelihood (ML) model implemented in the program PUZZLE v. 4.0.2 (30), indicated that none of the sequences included in our analyses significantly differed in nucleotide composition from this distribution. All maximum parsimony (MP) analyses were performed with PAUP* (31) with heuristic searches. We checked whether “simple”, “closest”, and “random” stepwise-addition sequences yielded identical trees. Other settings were: TBR branch-swapping, MULPARS, and zero-length branches collapsed. To avoid local optima, we performed 104 replicates both with random starting trees and with starting trees obtained by random stepwise addition. All characters were first weighted equally. Stability of the cladograms was tested with the Goloboff fit criterion (32) with k = 0, 2, 4, and 8. We estimated the reliability of the various inferred clades by bootstrapping (400 replicates) (33). By using the program TREEROT v.2 (34), we computed for all branches the Bremer support (BS)—the number of additional character transformations necessary to collapse an internal branch (35)—as an alternative to bootstrap values (BV) to estimate clade stability. We also performed topology-dependent PTP (T-PTP; ref. 36) analyses to statistically test monophyly of selected clades. Using a priori T-PTP is valid because it tests a hypothesis of monophyly formulated before our phylogenetic analyses (37). We compared unordered parsimony analyses with weighted-parsimony analyses. In the latter, substitution types for which saturation was obvious from saturation plots (cf. above) were excluded. All MP analyses were separately performed with each of the four gene fragments and with the combined (12S + 16S + COI + COII) data set. Partition homogeneity tests (ILD, ref. 38) implemented in PAUP* indicated that the protein-coding and ribosomal RNA genes were not significantly incongruent (P = 0.726). We used the ML method of phylogeny inference on a reduced data set with the following settings (PAUP*): empirical nucleotide frequencies, Ti/Tv ratio and proportion of invariable sites (Pinv) estimated by ML, HKY model (39) with rate heterogeneity, rates for variable sites assumed to follow a distribution (four categories) with shape parameter estimated by ML, and TBR branch-swapping. Given the high computation burden of ML analyses, it was practical to perform a bootstrap analysis (400 replicates) only by constraining ML parameters values (Pinv, Ti/Tv ratio, and γ shape) to those obtained in the ML search on the original (nonresampled) data set. ML analyses of the full data set (35 species) were performed after estimating the γ and Ti/Tv parameters from the strict consensus of the MP trees. Alternative phylogenetic hypotheses were compared statistically by means of Kishino–Hasegawa (KH) ML ratio tests (40). ML trees were also constructed by using the “quartet puzzling” heuristic (PUZZLE v.4.0.2; ref. 30) with Ti/Tv, nucleotide frequencies, and γ parameter estimated from the data, 10,000 puzzling steps, and HKY model. Minimum evolution (ME) and neighbor-joining (NJ) analyses were performed on the combined data set (including bootstrapping, 400 replicates) with PAUP* using LogDet distances (41), which have the advantage to be insensitive to variation of the substitution probability matrix, hence, to variations of base composition, throughout the tree. Sites estimated invariable by ML were removed before distance calculations because it makes the LogDet transform more robust to unequal substitution rates across sites (42).
Ecological Character Mapping.
Evolution of host-plant association and chemical defense was parsimoniously reconstructed with MACCLADE 3.05 (27). We used the minimum number of host shifts within the interrupta group required by the inferred phylogeny as a statistic. We treated host affiliation as an unordered binary character and compared the distributions of the minimum number of host-plant shifts inferred on all possible resolutions of the strict consensus of MP trees before and after 10,000 random permutations of the character states. A smaller observed value of shifts for the not-permuted data would suggest “historical inertia” (43), i.e., phylogenetic constraint, on the insect/plant associations.
Dating Evolutionary Events.
We estimated divergence time for various nodes on the phylogenetic trees by using a molecular clock and CO calibrations ranging from 0.8 × 10−8 to 1.7 × 10−8 substitution/site/year (44, 45). We tested whether a molecular clock could be rejected for the CO sequences under “two-cluster” and “branch length” tests (46) implemented in the LINTRE software.
Results
Phylogenetic Analyses.
Overall rates of divergence of COI and COII fragments are approximately twice as large as corresponding divergences in the 12S and 16S fragments. The unweighted MP heuristic analyses of the combined data set yield 120 MP trees whose strict consensus is shown in Fig. 1a. The monophyletic group interrupta (shaded box; Fig. 1) is strongly supported by bootstrapping (bootstrap value = 100), decay index (=25), and T-PTP analyses (P < 0.001). As 12S and 16S saturation plots indicate clear Ti saturation for pairwise distances comparisons >6%, we also performed weighted MP analyses (with Ti ignored or weighted three times less than transversions). The resulting trees are fully compatible with the strict consensus shown in Fig. 1a, although two nodes are collapsed within the interrupta group. All branches shown in Fig. 1a are stable to Goloboff weighting. COI and COII saturation plots do not show evidence of Ti saturation for any of the three codon positions. Analyses on the data set in which unstable aligned characters are excluded yield results basically identical to those described above.
ML analysis yields one tree (Fig. 1b) that is largely compatible with the MP analyses (Fig. 1a). Constraining nonmonophyly of the interrupta group requires a significant decrease in likelihood (KH test; ref. 40) both under exclusion (δln L = 73.37794, P < 10−4) and inclusion (δln L = 27.45876, P = 0.0288) of their sister species Chrysomela salicivorax. Hence, any tree not compatible with the monophyly of the interrupta group is significantly worse than the tree shown in Fig. 1b. ML analysis of a reduced data set (17 taxa) yields a single best tree (Fig. 2) with estimated Ti/Tv ratio, Pinv, and γ parameters equal to 1.73, 0.42, and 0.53, respectively. Constraining these parameter values for each of 400 bootstrap replicates yield BV indicated above the branches (Fig. 2). The ME and NJ analyses of the combined data set using LogDet distances (Pinv = 0.42) give topologies largely consistent with those shown in Fig. 1 a and b. None of the topology differences obtained through shifting among ML, MP, and distance approaches and/or excluding unstable aligned positions have any impact on the interpretation of host-affiliation and chemical defense evolution discussed below.
Ecological Character Mapping.
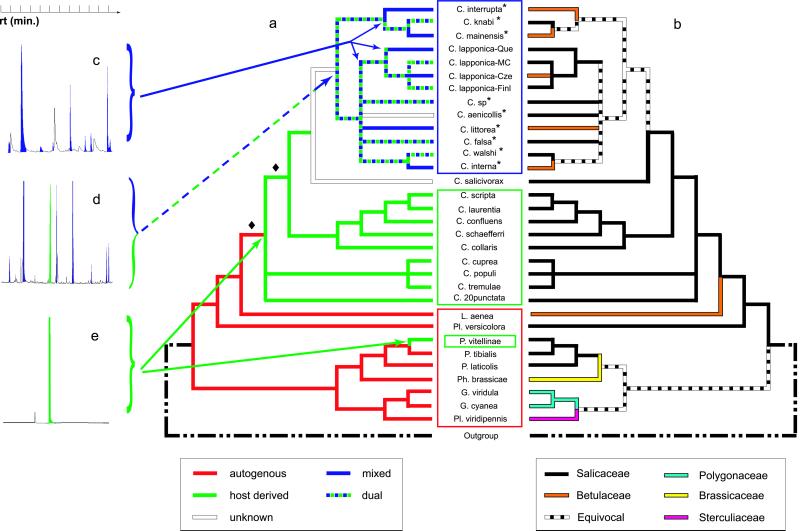
MP reconstruction of the evolution of Chrysomeline larvae chemical defense on any of the MP, ML, and optimal distance trees discussed above unambiguously indicate that de novo synthesis of iridoids was replaced by a host-derived salicylaldehyde strategy, which itself evolved into a mixed metabolism with synthesis of butyric acids (Fig. 3a). These analyses demonstrate that a switch from an autogenous to a fully host-dependent defense strategy did not prevent some more recent lineages (the interrupta group) to partially escape this dependence by evolving a defense strategy requiring a mixed plant/insect biosynthetic route. Although we were not able to investigate chemical data for C. salicivorax (we did not have access to live specimens), it is very likely that this species produces salicylaldehyde as do all Chrysomela species feeding on Salicaceae. However, the pattern of successive shifts in chemical defense suggested here (from autogenous to host-derived to mixed) would remain valid even in the eventuality C. salicivorax would produce butyric acids (mixed metabolism): the localization of the emergence of the mixed metabolism would simply be moved one node down the leaf beetle phylogeny, i.e., at the node grouping the interrupta group with C. salicivorax. MP reconstruction of ancestral host-plant affiliations (Fig. 3b) suggests a minimum of five independent host-plant shifts within the interrupta lineage. As this value falls within the distribution of host shifts after random permutations of the host-affiliation among taxa, it seems that host shifts within the interrupta group are not subjected to significant phylogenetic or chemical constraints. Even under the radical opinion that only the nodes supported by a 100% bootstrap value are inferred with high confidence, a minimum of three host shifts are still required under all resolutions of relationships within the interrupta clade. Obviously, the permutation test was performed in the context of the limited number of host-plant genera with which the interrupta lineage is associated, i.e., although there is no evidence of phylogenetic or chemical constraints restricting shifts between Salix/Populus and Alnus/Betula, there must be constraints to host shifts at a different level as the species of the interrupta clade only use these plant families, whereas others are potentially available (cf. below).
Figure 3.
(a) MP reconstruction of chemical defense strategies on the MP strict consensus from Fig. 1a; red, autogenous monoterpene iridoids; green, salicylaldehyde derived from salicin (sequestered from Salicaceae); blue, mixed metabolism, i.e., butyric acids and esters of them originating from esterification of de novo-synthesized butyric acids by alcohols taken up from the food plant. Some taxa within the interrupta lineage have a dual defense combining the mixed and host-derived metabolisms. (b) MP reconstruction of host plant associations. Que, Queyras (France); MC, Massif Central (France); Cze, Czech Republic; Finl, Finland. Asterisks indicate North American species within the interrupta group. (c–e) Typical gas chromatograms characterizing, mixed-metabolism, dual, and host-derived defenses, respectively; blue peaks, butyric acids and esters of them; green peak, salicylaldehyde; rt, retention time. The low bootstrap support for the two nodes indicated by a diamond do not challenge the general pattern of chemical defense evolution uncovered here (see text for details).
Dating Evolutionary Events.
Using all species from the interrupta clade as ingroup together with a chosen subset of outgroup taxa, COII molecular clock could not be rejected under either the “two-cluster” or the “branch length” tests. A linearized tree (i.e., with the assumption of rate constancy) was then constructed and nodes were dated by using CO rates. This yielded a date estimate for the most recent common ancestor of the interrupta group ranging from 1.1 to 2.3 million years ago (mya).
Discussion
Evolution of Chemical Characters.
As indicated above, phylogenetic reconstruction of the evolution of Chrysomelina larvae chemical defense unambiguously indicates an evolutionary sequence in which the autogenous synthesis of iridoid monoterpenes is the ancestral character state (color-coded in red in Fig. 3a), the host-plant derived strategy leading to the release of salicylaldehyde (green) being a secondary biogenetic route. The shift of chemical defense from autogenous to host-derived is energetically advantageous: in addition to economizing the biosynthesis of iridoids, larvae recover the glucose produced through catabolization of salicin into salicylaldehyde (47). This host-derived chemical defense based on sequestration of salicin convergently evolved in the species Phratora vitellinae (Fig. 3a; ref. 48). Given that the autogenous and host-derived defenses are quite dissimilar, the likelihood of such a convergent event might seem exceedingly low. However, chemical characterization of enzymes that are involved in the respective biogenetic routes indicates that the shift of defense might be because of a change in the specificity of an alcohol dehydrogenase (49, 50), allowing the use of salicin as a new substrate for the enzyme. Hence, the iridoid and salicin metabolisms are probably mutually exclusive (unless there were gene duplication followed by functional diversification). Two nodes (indicated by ♦ in Fig. 3a) relevant to the pattern of evolution of chemical defense are supported by low bootstrap values (54 and 42%) but are stable to Goloboff weighting. Still, the lineage leading to the species L. aenea could be conceivably moved one or two nodes up the tree into the Chrysomela clade. This would require the reversal (not observed elsewhere in the tree) from host-derived to autogenous chemical defense (from green to red, Fig. 3a). Even under each of the possible alternative topologies taking into account the low bootstrap support of the two nodes in question, chemical defense would still have evolved from an ancestral autogenous chemistry to a host-derived metabolism.
In the framework of the “dead-end” hypothesis (14–20), one could expect that the described shift in host plant and defense strategy had condemned the newly defined lineage to remain on Salicaceae throughout its existence, i.e., full dependence of the insect defensive strategy on salicin should have prevented any further shift toward non-Salicaceae host species. Our analyses conclusively demonstrate that this scenario is incorrect as far as the chrysomeline beetles are concerned. Indeed, the host-derived defense evolved into a mixed metabolism (blue in Fig. 3a), based on the de novo synthesis of butyric acids and their esterification with various alcohols taken up from the food plant. Given the high support for the monophyly of the interrupta group (see above), this most-derived strategy is a shared derived character state for the whole group. Because alcohols involved in the mixed metabolism are common components of plants (23), it can be considered less dependent than the salicylaldehyde biogenetic route on the specific host plant. Furthermore, our gas chromatography/mass spectrometry analyses of defensive secretions (51) demonstrate the plasticity of defense strategies within the interrupta group. Indeed, species exclusively found on Betulaceae secrete butyrates (mixed metabolism) and do not release any salicylaldehyde (Fig. 3c) as its precursor is not present in their food plants. On the other hand, species found on Salicaceae exhibit a “dual defense” as they not only produce butyric acids but also retain the ancestral ability to produce salicylaldehyde (Fig. 3d). Even if it was experimentally demonstrated that the salicin metabolism is not potentially functional in species of the interrupta group living on Betulaceae, the “dual defense” can only be the ancestral condition for the interrupta lineage. Indeed, the alternative hypothesis of multiple convergent evolution of the dual defense would require that, in the presence of the butyric acid metabolism, the loss of the ability to produce salicylaldehyde is more costly than its gain. In the unlikely eventuality, discussed above, L. aenea (producing iridoid monoterpenes) would be nested within the paraphyletic group of Chrysomela species with salicylaldehyde chemistry, our hypothesis of plasticity (here, through reversals) of defense strategies despite the existence of strong chemical constraints, would even be strengthened, albeit marginally.
Evolution of Host-Plant Affiliations.
As inference of the ancestral host plant for the interrupta group is ambiguous (Fig. 3b), identification of the dual defense adaptive value for the common ancestor of the interrupta group would be necessary for understanding the mode and tempo of host shifts within the lineage. More specifically, if dual defense provided a selective advantage per se (because, e.g., a larger diversity of chemical deterrents at the individual level could be adaptive against specialist predators), one could hypothesize that the new genetic variant which evolved “dual defense” remained on Salicaceae (where dual defense is effective) and shifts toward Betulaceae incidentally and convergently occurred in several lineages. On the other hand, if the key parameter in the evolution of dual defense was that it provided disruption of the mandatory association with plants containing salicin, it is likely that it is a shift toward Betulaceae which insured survival of the genetic variant (because it could, e.g., lower intraspecific competition or would correspond to the selection of an enemy-free space). Under that latter scenario, multiple reversals to Salicaceae hosts have been only possible for chrysomeline species that maintained the ability to metabolize salicin into salicylaldehyde. Obviously, these two possibilities are extreme schemes bordering a range of intermediate hypotheses characterized by a combination of reversals and convergent events. Whichever was their exact dynamic, host shifts were always followed by a retention of oligophagy. Identification of the major constraint(s) (e.g., developmental, behavioral, ecological) involved in this striking pattern would require further research beyond the scope of molecular phylogenetic investigations.
As their butyric acid-based protection allows the species of the interrupta group to escape from the association with Salicaceae, one can wonder why these insects are pledged exclusively to plant species of the genera Salix and Populus in the family Salicaceae, or to genera Alnus and Betula in the family Betulaceae. Indeed, one would expect the newly derived chemistry to allow host shifts toward other plant families. The combination of plant ecological data with our molecular phylogenetic analyses indicates that host availability might have been the key parameter for this pattern to appear. Indeed, the holarctic Chrysomela species belonging to the interrupta lineage currently all have similar biotopes (humid meadows and riverbanks) in which host shifts could occur by opportunistic explorations among Salicaceae and Betulaceae species. Furthermore, phytopaleontological data from late quaternary Northwest Canada (52), Alaska (53), Siberia (54), and Northwest Europe (55) indicate that species from the genera Salix/Populus and Alnus/Betula were then the most abundant arborescent plants and were closely associated into boreal shrub tundra and open forests.
As the interrupta lineage includes both North-American (indicated by asterisks in Fig. 3) and European species, and our molecular estimate for the origin of that group ranges from 1.1 to 2.3 mya, members of it must have crossed the Bering Strait after its geological opening (>4.8 mya). The period at which the interrupta group originated is characterized by a series of glaciations and warmer periods. As invasions of vegetation that followed the retreat of glaciers progressively led to a mixed Salicaceae–Betulaceae forest (56), the association of chrysomelines from the interrupta lineage with host plant species belonging exclusively to these two families is not surprising, even though they are neither chemically nor phylogenetically closely related (57, 58).
Conclusions
The most ancestral defense strategy of the Chrysomelina beetles we investigated is based on an autogenous metabolism producing monoterpene iridoids. Two lineages of this group convergently specialized by developing an energetically less costly host-derived metabolism based on the production of salicylaldehyde from salicin, a secondary plant compound characteristic of Salicaceae. As this evolutionary transition probably required mutation of a single enzyme, it is likely that the iridoid and salicin metabolisms are mutually exclusive. Our phylogenetic analyses further indicate that, although development of the salicin metabolism makes the insect tightly dependent on the chemistry of the host plant (Salicaceae), it did not prevent a lineage from escaping this subordination through the development of a yet more derived mixed metabolism, potentially compatible with a large number of new host-plant associations. Incidentally, the individuals that experienced these shifts probably also overcame additional chemical constraints because specific plant compounds can act both as stimuli for feeding/oviposition and as deterrents/toxins (59). Our analyses provide an example of a mechanistic explanation for a deviation from the dead end hypothesis. Furthermore, as the mixed and salicylaldehyde-based defense strategies involve independent chemistries, the development of a dual defense (with both salicin and mixed metabolisms) was made possible. Because we feel that independent losses of the salicin metabolism are more likely than their multiple independent gains, we view the dual defense as ancestral for the whole interrupta group. Still, only careful characterization of the relative selective advantages provided respectively by the dual metabolism per se and by the loss of dependence to Salicaceae could conceivably allow to uncover the convergent/reversal events of host shifts between Salicaceae and Betulaceae during the evolution of host affiliation within the interrupta clade. Finally, association of members of the interrupta group either with Salicaceae or Betulaceae, and not with other plant families, can be explained by an historical constrain: phytopaleontological data suggest that the host-plant genera in these families dominated shrub tundra and open forest habitats when the interrupta group originated and diversified.
Acknowledgments
We thank P. Mardulyn, E. Neuvonen, L. LeSage, Y. P. Yu, and V. Krischik for providing samples, and F. Damblon for helping in the interpretation of paleontological data. A.Termonia is supported by a Ph.D. grant from the “Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture (FRIA). J.M.P. is supported by grants of the Belgian Fund for Joint Basic Research (2.4505.98). M. C. Milinkovitch's Unit of Evolutionary Genetics is supported by grants from the National Fund for Scientific Research Belgium (FNRS), the Free University of Brussels (ULB), the ”Communauté Française de Belgique“ (ARC 98/03-223), the Defay Fund, and the Van Buuren Fund.
Abbreviations
- COI
cytochrome oxidase I
- Tv
transversion
- Ti
transition
- MP
maximum parsimony
- ML
maximum likelihood
- mya
million years ago
Footnotes
References
- 1.Mitter C, Farrell B, Futuyma D J. TREE. 1991;6:290–293. doi: 10.1016/0169-5347(91)90007-K. [DOI] [PubMed] [Google Scholar]
- 2.Farrell B D. Science. 1998;281:555–558. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- 3.Wilf P, Labandeira C C, Kress W J, Staines C L, Windsor D M, Allen A L, Johnson K R. Science. 2000;289:291–294. doi: 10.1126/science.289.5477.291. [DOI] [PubMed] [Google Scholar]
- 4.Ehrlich P R, Raven P H. Evolution. 1964;18:586–608. [Google Scholar]
- 5.Menken S B J, Roessingh P. In: Endless Forms Species and Speciation. Howard D J, Berlocher S H, editors. Oxford: Oxford Univ. Press; 1998. pp. pp.145–156. [Google Scholar]
- 6.Futuyma D J, McCafferty S S. Evolution. 1990;44:1885–1913. doi: 10.1111/j.1558-5646.1990.tb04298.x. [DOI] [PubMed] [Google Scholar]
- 7.Becerra J X. Science. 1997;276:253–256. doi: 10.1126/science.276.5310.253. [DOI] [PubMed] [Google Scholar]
- 8.Farrell B, Mitter C. Evolution. 1990;44:1389–1403. doi: 10.1111/j.1558-5646.1990.tb03834.x. [DOI] [PubMed] [Google Scholar]
- 9.Mardulyn P, Milinkovitch M C, Pasteels J M. Sys Bio. 1997;46:722–747. doi: 10.1093/sysbio/46.4.722. [DOI] [PubMed] [Google Scholar]
- 10.Thompson J N. The Coevolutionary Process. Chicago: Univ. of Chicago Press; 1994. [Google Scholar]
- 11.Strong D R, Lawton J H, Southwood R. Insects on Plants. Cambridge, MA: Harvard Univ. Press; 1984. [Google Scholar]
- 12.Bernays E, Graham M. Ecology. 1988;69:886–892. [Google Scholar]
- 13.Rowell-Rahier M, Pasteels J M. Herbivores: Their Interactions with Secondary Plant Metabolites. Vol. 2. San. Diego: Academic; 1992. pp. 244–277. [Google Scholar]
- 14.Simpson G G. The Major Features of Evolution. New York: Columbia Univ. Press; 1953. [Google Scholar]
- 15.Mayr E. Animal Species and Evolution. Cambridge, MA: Harvard Univ. Press; 1963. [Google Scholar]
- 16.Feeny P. In: Coevolution of Animals and Plants. Gilbert L E, Raven P H, editors. Austin, TX: Univ. of Texas Press; 1975. p. 246. [Google Scholar]
- 17.Futuyma D J, Moreno G. Annu Rev Ecol Syst. 1988;19:207–233. [Google Scholar]
- 18.Moran N. Am Nat. 1988;132:671–706. [Google Scholar]
- 19.Wiegmann B M, Mitter C, Farrell B. Am. Nat. 1993. 142. [Google Scholar]
- 20.Kelley S T, Farrell B D. Evolution. 1998;52:1731–1743. doi: 10.1111/j.1558-5646.1998.tb02253.x. [DOI] [PubMed] [Google Scholar]
- 21.Veith M, Lorenz M, Boland W, Simon H, Dettner K. Tetrahedron. 1994;50:6859–6874. [Google Scholar]
- 22.Pasteels J M, Grégoire J-C, Rowell-Rahier M. Annu Rev Entomol. 1983;28:263–289. [Google Scholar]
- 23.Schulz S, Gross J, Hilker M. Tetrahedron. 1997;53:9203–9212. [Google Scholar]
- 24.Simon C, Frati F, Beckenbach A, et al. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- 25.Hsiao T H, Pasteels J M. Advances in Chrysomelidae Biology. Vol. 1. Leiden: Backhuys; 1999. pp. 321–342. [Google Scholar]
- 26.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddison W P, Maddison D R. MacClade. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 28.Loytynoja, A. & Milinkovitch, M. (2001) Bioinformatics, in press.
- 29.Gatesy J, DeSalle R, Wheeler W. Mol Phylogenet Evol. 1993;2:152–157. doi: 10.1006/mpev.1993.1015. [DOI] [PubMed] [Google Scholar]
- 30.Strimmer K, Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 31.Swofford D L. paup*: Phylogenetic Analysis Using Parsimony (and Other Methods) Sunderland, MA: Sinauer; 2000. , Version 4.0b4a. [Google Scholar]
- 32.Goloboff P A. Cladistics. 1993;9:83–91. doi: 10.1111/j.1096-0031.1993.tb00209.x. [DOI] [PubMed] [Google Scholar]
- 33.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 34.Sorenson M D. treerot. Boston: Boston University; 1999. , Version 2.0. [Google Scholar]
- 35.Bremer K. Cladistics. 1994;10:295–304. [Google Scholar]
- 36.Faith D P. Syst Zool. 1991;40:366–375. [Google Scholar]
- 37.Faith D P, Trueman J W H. Syst Biol. 1996;45:580–586. [Google Scholar]
- 38.Farris J S, Källersjö M, Kluge A G, Bult C. Cladistics. 1994;10:315–319. [Google Scholar]
- 39.Hasegawa M Y, Kishino H, Yano T. J Mol Evol. 1985b;21:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 40.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 41.Lockhart P J, Steel M A, Hendy M D, Penny D. Mol Biol Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- 42.Waddell P J. Ph.D. dissertation. Massey, New Zealand: Massey University; 1995. [Google Scholar]
- 43.Maddison W P, Slatkin M. Evolution. 1991;45:1184–1197. doi: 10.1111/j.1558-5646.1991.tb04385.x. [DOI] [PubMed] [Google Scholar]
- 44.Gómez-Zurita J, Juan C, Petitpierre E. Mol Phylogenet Evol. 2000;14:304–317. doi: 10.1006/mpev.1999.0712. [DOI] [PubMed] [Google Scholar]
- 45.Funk D J, Futuyma D J, Ortí G, Meyer A. Evolution. 1995;49:1008–1017. doi: 10.1111/j.1558-5646.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- 46.Takezaki N, Rzhetsky A, Nei M. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 47.Rowell-Rahier M, Pasteels J. J Chem Ecol. 1986;12:1189–1203. doi: 10.1007/BF01639004. [DOI] [PubMed] [Google Scholar]
- 48.Pasteels J, Rowell-Rahier M, Braekman J, Daloze D. Biochem Sys Ecol. 1984;12:395–406. [Google Scholar]
- 49.Pasteels J M, Duffey S, Rowell-Rahier M. J Chem Ecol. 1990;16:211–222. doi: 10.1007/BF01021280. [DOI] [PubMed] [Google Scholar]
- 50.Veith M, Oldham N, Dettner K, Pasteels J, Boland W. J Chem Ecol. 1997;23:429–443. [Google Scholar]
- 51.Termonia A, Pasteels J M. Chemoecology. 1999;9:13–23. [Google Scholar]
- 52.Szeicz J M, MacDonald G M, Duk-Rodkin A. Paleobiology. 1994;113:351–371. [Google Scholar]
- 53.Ellias S A, Short S K, Waythomas C F. Arctic. 1996;49:292–305. [Google Scholar]
- 54.Ukraintseva V V. In: Siberia. Agenbroad L D, Mead J I, Hevly R H, editors. South Dakota: Hot Springs; 1993. p. 309. [Google Scholar]
- 55.Paus A. Rev Paleobot Palynol. 1995;85:243–262. [Google Scholar]
- 56.Cox C B, Moore P D. Biogeography. An Ecological and Evolutionary Approach. Oxford: Blackwell Scientific; 1993. [Google Scholar]
- 57.Hegnauer R. Chemotaxonomie der Pflanzen. Vol. 6. Basel: Birkhäuser; 1973. pp. 240–258. [Google Scholar]
- 58.Savolainen V, Chase M W, Hoot S B, Morton C M, Soltis D E, Bayer C, Fay M F, Vruijn A Y d, Sullivan S, Qiu Y-L. Syst Biol. 2000;49:306–362. doi: 10.1093/sysbio/49.2.306. [DOI] [PubMed] [Google Scholar]
- 59.Bernays E A, Chapman R F. In: Host-Plant Selection by Phytophagous Insects. Miller T A, Van Emden S H, editors. New York: Chapman & Hall; 1994. [Google Scholar]