Abstract
The phytohormone cytokinin is a key player in many developmental processes and in the response of plants to biotic and abiotic stress. The cytokinin signal is perceived and transduced via a multistep variant of the bacterial two-component signaling system. Most of the research on cytokinin signaling has been done in the model plant Arabidopsis thaliana. Research on cytokinin signaling has expanded to a much broader range of plants species in recent years. This is due to the natural limitation of Arabidopsis as a model species for the investigation of processes like nodulation or wood formation. The rapidly increasing number of sequenced plant genomes also facilitates the use of other species in this line of research. This review summarizes what is known about the cytokinin signaling in the different plant species and highlights differences to Arabidopsis.
Key words: cytokinin, signaling, cytokinin signal transduction, cytokinin receptor, histidine phosphotransfer protein, response regulator, two-component system
Phytohormones are well known regulators of plant development.1 In addition, they also play an important role in mediating the response of plants to biotic and abiotic factors in the environment. Cytokinins, N6-substituted adenine derivates, are one class of phytohormones.2 The cytokinin signal is perceived and transduced via a multi-step variant of the two-component system (TCS). While the TCS is the principal signaling system in bacteria, among higher eukaryotes it is found only in plants.3
The current model of cytokinin signaling predicts that cytokinin is perceived by a membrane bound hybrid histidine kinase receptor (HK). The binding of the ligand leads to an autophosphorylation of the receptor and is followed by an intramolecular transfer of the phosphoryl residue to the receiver domain of the receptor. Subsequently, the phosphate is transferred to a histidine phosphotransfer protein (HPt), which translocates to the nucleus where it activates type-B response regulators (type-B RRs). This class of response regulators are Myb-type transcription factors and among their target genes are the type-A response regulators (type-A RRs). The type-A RRs act as negative feedback regulators of the cytokinin signaling pathway.4
Recently, phylogenetic analysis aimed at unraveling the origin of this signaling pathway identified the moss Physcomitrella patens as the most early diverging organism examined to encode members of all four protein families in its genome. While the analysis of the also investigated algal species revealed the presence of genes for HPts and type-B RRs, no sequences encoding for putative cytokinin receptors or type-A RRs were detected.5
Most of the research on cytokinin signaling to date has been carried out in the model plant Arabidopsis thaliana. However, studies in other plants have not only contributed to the elucidation of this pathway but also highlighted the cross-talk between cytokinin signaling and other non-hormone signaling pathways, i.e., signaling pathways regulating nitrogen and sulfur metabolism,6–9 nodulation,10–14 pathogen defense,15 heavy metal response16 and drought stress.17,18
The function of cytokinin signaling genes are investigated in two types of experiments. Single genes of the cytokinin signaling pathway were identified in screens connecting cytokinin signaling to the respective stimuli. Systematic approaches characterizing all family members of the cytokinin signaling pathway in a given species were conducted in species with completely sequenced genomes. Examples for such methodical experiments can be found in rice, poplar, lotus and grape.7,19–24 With the rapidly increasing number of completely sequenced plant genomes and the new possibilities offered by the arrival of the next generation sequencing techniques for transcriptome analysis, the opportunity for a larger number of targeted analysis in plant species other than Arabidopsis thaliana is growing dramatically. This is not only welcome, but also necessary as the influence or crosstalk of cytokinin in many biological processes, such as nodulation, wood formation and pathogen response to species not affecting Arabidopsis cannot at all or can only rudimentarily be investigated in Arabidopsis. Thus cytokinin research is bound to profit greatly from a broadening in focus to include diverse plant species.
The aim of this review is to summarize the current state of knowledge about cytokinin signaling in different plant species beyond Arabidopsis thaliana (for a comprehensive overview see Sup. Table 1). In the following sections we will review what is known for members of each of the four protein families involved in the cytokinin signaling pathway. For a detailed description of cytokinin signaling in Arabidopsis see recent reviews.25–27
Cytokinin Receptors
One of the processes where cytokinin plays a crucial role but cannot be investigated in Arabidopsis is root nodule formation. Three publications describe the role of a cytokinin receptor from lotus, Lotus Histidine Kinase 1 (LHK1), in this symbiotic process.10,12,14 Receptor loss-of-function mutants failed to respond to the rhizobial signal and did not perform the first step of nodule formation—the division of cortical cells.12 In contrast, a constitutively active mutant displayed spontaneous development of nodules even in the absence of the symbiotic rhizobial bacteria or their signaling molecules.14 This spontaneous nodulation can be suppressed by addition of abscisic acid (ABA).10 Expression analysis detected receptor transcripts in uninfected root tissues as well as in nodules. Heterologous complementation of bacterial and yeast mutants further confirmed LHK1 function as a cytokinin receptor.12,14 Medicago truncatula is also a root nodule forming species containing three cytokinin receptors. An RNAi approach indicated that at least one of the putative cytokinin receptors also acts in nodule formation.11 Experimental data showing expression of the histidine kinase genes in newly formed nodules supported this assessment.28,29 Further expression analysis indicated that at least two of the cytokinin receptors in Medicago might play a role in abiotic stress response as well.30
Another aspect of plant biology where Arabidopsis cannot serve as a model is wood formation and the influence that cytokinin has on this developmental process. With the completed genomic sequence at hand, poplar is often used as a model organism for tree biology. As cambium is the tissue which gives rise to wood and cytokinin plays a decisive role in cambium development,31 putative cytokinin receptors of poplar have been investigated in this context.32 For four of the five putative cytokinin receptors of poplar, expression was detected in the cambium. The same group investigated the expression pattern of a putative cytokinin receptor from birch and found expression in the cambial zone as well. Several experiments including analysis of grafts, callus formation and stem diameter using cytokinin deficient transgenic poplar plants clearly demonstrated the role of cytokinin as a major regulator of cambium development.32
Other functional studies of cytokinin receptors have been focused on their core role in cytokinin signaling itself. In 2004, Yonekura-Sakakibara and colleagues investigated the expression patterns of three cytokinin receptors of corn in various tissues and while they found large overlaps in the temporal and spatial expression patterns of the three receptors, they also detected tissue specificity of the receptor transcripts.33,34 The cytokinin binding of the maize receptors revealed specificities similar to those detected in the respective homologs in Arabidopsis.35,36 Data describing putative cytokinin receptors in other plant species are limited to transcriptome analysis. Experiments in rice detected transcripts of putative cytokinin receptors in all tissues investigated.19,21,37 At least in one case the transcription level of the receptors increased in response to salt stress and dehydration, indicating a role for cytokinin in response to abiotic stress.17,18 Northern blot analysis in Catharanthus roseus showed a strong expression of a putative cytokinin receptor into several organs, but mainly in the flower.38 Expression of homologs of the Arabidopsis cytokinin receptors were also detected in lupine,39 tomato,40 potato15 and in grape.7 The main features of the cytokinin receptors across the different species have been compiled in Table 1.
Table 1.
Compilation of the function associated with cytokinin receptors from different species
| Species | Involved in /expression changed upon treatment with | ||
| Betula pendula BpCre1; BpHK2; BpHK3 | pWOL::BpCre1 complemented Arabidopsis triple receptor mutant32 | ||
| Brassica juncea BjCre1 | Downregulation by arsenite16 | ||
| Catharanthus roseus CrCKR1 | Downregulation by auxin, JA, NaCl, cytokinin (long-term)38 | ||
| Lotus japonicas LHK1; LHK2 | Nodulation (LHK1), LHK1 point mutations caused differences in nodulation10,12,14 | ||
| Lupinus albus LaHK1 | Upregulation upon dark stress39 | ||
| Lycopersicon esculentum CRK | Expression varies with growth conditions (subsurface)40 | ||
| Medicago sativa MsHK1 | Upregulation by salt stress and dark stress28,39 | ||
| Medicago trunculata MtCRE1; MtHK2; MtHK3 | Expression detected11; upregulation by salt stress and cytokinin (MtCRE1)11; MtHK2 expression upregulated by salt stress31; MtCRE1-RNAi mutant showed no phenotypical alterations but less nodules upon treatment with S. meliloti11; MtCRE1-expression is upregulated by treatment with S. meliloti29 | ||
| Oryza sativa OsHK1; OsHK2; OsHK3; OsHK3a; OsHK3b; OsHK4; OsHK4a; OsHK4b, Os HK5; OsHK6; OsETR4 | Expression detected18,19,21,37 Upregulation by salt stress, dehydration, cold stress (OsHK3)17 | ||
| Populus trichocarpa PtCRE1a; PtCRE1b; PtHK1; PtHK2; PtHK3a; PtHK3b | Expression detected32 Upregulation by PEG (PtHK1)47; Interaction of PtHK1 with PtHPT2 in yeast two-hybrid system47 | ||
| Solanum sparsipilum SsCRE1 | Expression detected15 | ||
| Vitis vinifera VvCyt1; VvCyt2; VvCyt3 | Downregulation by sulfur depletion plus cytokinin (VvCyt1, VvCyt2)7 | ||
| Zea maize ZmHK1, ZmHK2; ZmHK3a; ZmHK3b | Dose-dependent activity upon cytokinin treatment in E. coli (except ZmHK3b); best ligands were iP for ZmHK1; tZ and tZR for ZmHK2; iP and tZ for ZmHK3a34; expressed in seeds (ZmHK1, ZmHK2, ZmHK3a)33 |
Histidine Phosphotransfer Proteins
For most HPt proteins the focus of research has been on their role within the cytokinin signaling pathway. Several lines of evidence suggest a conserved role of HPts in maize and Arabidopsis. Transiently expressed ZmHP1-GFP localized to both the cytosol and the nucleus, as one would predict from the current model for cytokinin signaling.41 Phosphorelay and yeast two-hybrid experiments showed that at least ZmHP1, ZmHP2 and ZmHP3 are interacting with ZmRRs of both type-A and type-B families and that this interaction mediates a phosphotransfer.41 In addition, the maize ZmHP2 is the only full-length protein of the cytokinin signaling pathway in any species for which the three dimensional structure has been solved. This structure revealed that all the amino acids, which are conserved in plant HPts, surround the canonical histidine residue—probably forming a docking interface for the receiver domain of the receptors and response regulators.42 For ZmHP1, crystals and preliminary X-ray spectra were also reported, but thus far no crystal structure has been published.43
Functional analysis of HPts was also carried out in Catharanthus roseus. In cell-lines transformed with a CrHP1 RNAi construct induction of the type-A RR CrRR1 transcript was reduced, while the expression of the type-B RR CrRR5 remained unchanged by cytokinin treatment.13 The expression of CrHP1 itself in wild-type cell culture lines was not responsive to cytokinin. However, the addition of jasmonic acid or auxin led to an increase in CrHP1 expression, indicating a regulatory crosstalk between the signaling pathways of these phytohormones.13
In Medicago one of the HPts, MtHP2, was found to be upregulated upon salt stress, further corroborating the link between cytokinin and abiotic stress signaling detected in Arabidopsis.44,45 Transcripts of the HPt genes were detected in additional species, namely in orange,46 rice,18,19,21,37 poplar,47 wheat48 and in grape.7 The main features of the histidine phosphotransfer proteins across the different species have been compiled in Table 2.
Table 2.
Compilation of the function associated with phosphotransfer proteins from different species
| Species | Involved in /expression changed upon treatment with |
| Catharanthus roseus CrHPT1 | Upregulation by JA, downregulation by ethepon; RNAi-line showed decreased growth rate, lower expression of CrRR1 but not CrRR5 upon cytokinin treatment64 |
| Citrus sinensis CsHPT1 | Expression detected46 |
| Medicago trunculata MtHP2 | Upregulation by salt stress44 |
| Oryza sativa OsHP1; OsHP2; OsHP3, OsHP4; OsHP5 | Expression detected19,3,7; (OsHP1)18; (OsHP1 and OsHP2)21 |
| Populus trichocarpa PtHPT1, PtHPT2, PtHPT3, PtHPT4 | Expression detected47 |
| Triticum aestivum TaHP1, TaHP2, TaHP3 | Upregulation by 1µM BA, not tZ48 |
| Vitis vinifera VvHP1, VvHP2, VvHP3, VvHP | Upregulation by Phytoplasma bacteria (VvHP)66; regulated in sulfur starvation and cytokinin treatment (VvHP2, VvHP3)7 |
| Zea maize ZmHP1, ZmHP2, ZmHP3 | ZmHP1 and ZmHP3 localized to the nucleus and cytoplasm41; ZmHP2crystal structure solved42; no regulation by cytokinin ZmHP1 and ZmHP341, ZmHP256, or by nitrogen starvation (ZmHP2)56; yeast two-hybrid approach showed interaction of ZmHP3 with ZmRR1 and ZmRR1, of ZmHP1 with ZmRR1,8,9,10 and of ZmHP2 with ZmRR9, 1041; phosphorelay from ZmHP1 to ZmRR1, ZmRR8 and ZmRR4, ZmHP2 to ZmRR941; phosphorelay from ZmHP2 to ZmRR1 and ZmRR256 |
Type-B Response Regulators
In Medicago truncatula not only the cytokinin receptors were indicated in the nodule formation as described above, but also one of the three type-B RRs, MtRR1. This type-B RR was strongly induced in the earlier stages of nodule formation. However, MtRR1 RNAi plants did not display an altered nodulation phenotype and further research is needed to clarify the role of MtRR1 in this process.11
One of the poplar type-B RRs, PtRR13, was identified in a microarray experiment to be involved in adventitious root development. This finding was later confirmed by the phenotypes displayed by PtRR13 RNAi and transgenic dominant negative (ΔDDKPtRR13) plants, which showed defects in this process.24 Analysis of the expression patterns of all type-B RRs from poplar showed for at least three of the eleven type-B RRs a clear induction of gene expression after cytokinin treatment.23 This is surprising as according to the current model of cytokinin signaling, the type-A RRs are the only component of the pathway being transcriptionally regulated by cytokinin treatment and numerous microarray experiments from Arabidopsis confirmed this hypothesis.26,49,50
In maize, Asakura and colleagues showed that the three analyzed type-B RRs, ZmRR8, ZmRR9 and ZmRR10 can interact with the three ZmHPs in a yeast two-hybrid assay. These interactions were also verified in phosphorelay experiments for ZmRR8 and ZmRR9.41 Subcellular localization showed ZmRR8 to be located in the nucleus. The expression of all three type-B ZmRRs was not affected by cytokinin treatment.41
Taking advantage of the complete genome sequence, two groups analyzed the expression patterns of various TCS components in rice and could show differential expression patterns for the type-B OsRRs in various tissues, developmental stages and in response to abiotic stresses.17,19 The main features of the type-B RRs across the different species have been compiled in Table 3.
Table 3.
Compilation of the function associated with type-B response regtulators from different species
| Species | Involved in /expression changed upon treatment with |
| Catharanthus roseus CrRR5 | Expression detected64; Not affected by CrHPt1 antisense line13 |
| Lotus japonicas LjRRb1, LjRRb2, LjRRb3, LjRRb4, LjRRb5, LjRRb6, LjRRb7, LjRRb8, LjRRb9, LjRRb10, LjRRb11 | Expression detected20 |
| Medicago trunculata MtRR1, MtRR2 | Not cytokinin regulated11; MtRR1 is nod-factor dependent11, involved in nodulation11 and in developing seeds67 |
| Oryza sativa OsRR16, OsRR17, OsRR18, OsRR19, OsRR20, OsRR21, OsRR29, OsRR30/EDH1, OsRR33 | Not upregulated by cytokinin; expression level not changed by overexpression of OsRR3 and OsRR5 (OsRR16, OsRR17, OsRR18, OsRR20, OsRR21)60; slight regulation of OsRR16 by salt stress, cold stress and dehydration17, expression detected (OsRR16–21)19 |
| Populus trichocarpa PtRR12, PtRR13, PtRR14, PtRR15, PtRR16, PtRR17, PtRR18, PtRR19, PtRR20, PtRR21, PtRR22 | PtRR13 overexpressor and RNAi-line did not show any phenotype; constitutively active PtRR13 led to shorter and fewer roots, less adventious roots, callus formation at the cut site, greater tissue growth in absence of cytokinin23,24 |
| Zea maize ZmRR8, ZmRR9, ZmRR10 | ZmRR8 localized in the nucleus; Asp67/Asp78 important for ZmRR8/ZmRR9 to be phosphorylated by ZmHP1 and ZmHP2 in vitro; ZmRR8 receiver domain interacted strongly with ZmHP3, ZmRR with ZmHP2 and ZmRR10 with ZmHP141 |
| Vitis vinifera VvRRb1, VvRRb2, VvRRb3, VvRRb4, VvRRb5, VvRRb6 | Unaffected by sulfur depletion7 |
Type-A Response Regulators
Research on type-A RRs in maize points to roles in phyllotaxis and seed development. Aberrant phyllotaxy1 (abphyl1; also Zmrr3) was shown to be a regulator of embryo morphogenesis and shoot phyllotaxis.51 The ABPHYL1 expression was localized to the embryo and shoot apex.52 Detailed analysis of the mutant concluded that the phenotype was caused by reduced auxin levels leading to a larger shoot apical meristem, a delayed leaf initiation and altered leaf phyllotaxy.53 The type-A RRs ZmTcRR1 and ZmTcRR2 were reported to be specifically expressed in the transfer cell layer of developing maize kernels; however, so far no specific function was assigned to these genes.33,54 Other members of type-A RRs of maize were characterized for their role in cytokinin signaling. On the protein level, type-A RRs were shown to interact with ZmHPs in yeast two-hybrid and phosphorelay experiments. These in vitro experiments showed also that type-A ZmRRs auto-dephoshorylated much faster than type-B ZmRRs,41 which might be an important trait for their function as negative regulators of the cytokinin signaling pathway. Three type-A RRs, ZmRR1, ZmRR2 and ZmRR3, were shown to localize to the cytosol and the nucleus, while ZmRR4, ZmRR5 and ZmRR6 were found exclusively in the nucleus of transiently transformed onion epidermis cells.41 The transcription of all type-A ZmRRs expressed in the leaf blade was shown to be strongly induced by cytokinin.6,41,55 ZmRR1 and ZmRR2 were found to be involved in the nitrogen signal transduction,8,9,56,57 a node of crosstalk which has also been discovered in Arabidopsis.58
Experimental characterization of the type-A RRs of rice further emphasized the role of this protein family in plant development. Calli overexpressing OsRR6 were severely retarded in shoot regeneration and transgenic plants displayed a general dwarf phenotype with a poorly developed root system—consistent with a role of OsRR6 as a negative regulator of cytokinin signaling.59 Rice plants overexpressing either OsRR3 or OsRR5 exhibited a lower cytokinin sensitivity in root elongation and callus growth assays. Furthermore, in these transgenic plants other type-A RRs such as OsRR1, OsRR7, OsRR14 and OsRR15 were downregulated, while other members of the same gene family showed a higher level of transcription than in the wild type control.60 Also type-A RRs from other species were implicated in plant development. A recent study hints at a role for type-A RR, PipiRR1, during caulogenic induction in Pinus pinea. The respective transcript was detected in the cotyledons and increased after treatment with cytokinin in a dose-dependent manner.61
Type-A RRs of other species were shown to react to abiotic stimuli. The transcript level of PvRR1 of Phaseolus vulgaris increased during starvation experiments related to macronutrients such as phosphorus, potassium and nitrogen as well as upon addition of cytokinin, while it decreased after resupply of the nutrients.62 Two type-A RRs of Medicago, MtRR4 and MtRR5, were upregulated in response to salt stress, but also during nodulation.30 In contrast, the three type-A RRs of Catharanthus roseus seemed to be specifically induced by cytokinin.13,63,64
Systematic expression studies for all type-A RRs of a given species have been done in rice, lotus and poplar. The extensive analysis of the complete set of type-A RRs in rice revealed different temporal and spatial expression patterns of this class of OsRRs. The transcript level of most type-A OsRRs increased upon cytokinin treatment.19,21,22 Under different abiotic stress treatments, the expression of the type-A OsRRs was specifically and differentially up or downregulated.17 In Lotus japonicus, the expression of six of the seven tested type-A RRs was induced by cytokinin. For the seventh gene no transcript could be detected and the authors suspected it to be a non-functional gene as parts of the C-terminus are missing.20 In a similar experiment, investigating the members of this gene family in poplar, seven of the eleven type-A PtRRs were found to be induced by cytokinin.23 For PtRR7, a strong expression was found in the cambium. In plants with a lower cytokinin status due to the ectopic expression of AtCKX2, the expression level of PtRR7 was clearly reduced.32 The main features of the type-A RRs across the different species have been compiled in Table 4.
Table 4.
Compilation of the function associated with type-A response regulators from different species
| Species | Involved in /expression changed upon treatment with |
| Catharanthus roseus CrRR1, CrRR2, CrRR3 | Upregulation by cytokinin, but not auxin, salt stress or ABA; induction was inhibited by blocking of the receptors (CrRR1)64; CrRR1 expression downregulated in CrHP1-antisense plants13; not transcript detected for CrRR2 but for CrRR313 |
| Lotus japonicas LjRRa1, LjRRa2, LjRRa3, LjRRa4, LjRRa5, LjRRa6, LjRRa7 | Upregulated by cytokinin20; (LjRRa5)63 |
| Medicago trunculata MtRR3, MtRR4, MtRR5 | Regulated by cytokinin11, upregulated by symbiosis (MtRR4)29; partially nod-factor-dependent (MtRR4 is dependent, MtRR5 not)11; upregulated in MtCRE1-RNAi-roots29 |
| Oryza sativa OsRR1, OsRR2, OsRR3, OsRR4, OsRR5, OsRR6, OsRR7, OsRR8, OsRR9, OsRR10, OsRR11, OsRR12, OsRR13, OsRR14, OsRR15, OsRR41 | Expression detected17,19,21,22,60; upregulated by cytokinin (OsRR1–11, 14)21,22,60; partially regulated by dehydration, salt stress and cold stress17, overexpressors of OsRR3 and OsRR5 with more lateral roots and longer roots on cytokinin, but more sensitive towards cytokinin in callus formation and chlorophyll content assay; several type-ARRs downregulated in overexpressors of OsRR3 and OsRR560; overexpressed GFP fusion with OsRR6 shows accumulation in cytosol; overexpressor of OsRR6 has a dwarfed, less branched phenotype, a less developed root system and is sterile59; Osrr9/Osrr10 knockout plants are dwarfed and sterile22 |
| Phaseolus vulgaris PvRR1 | Upregulation by cytokinin, and depletion of K, N and P; localized to the nucleus62 |
| Pinus pinea PipiRR1 | Upregulation by cytokinin61 |
| Populus trichocarpa PtRR1, PtRR2, PtRR3, PtRR4, PtRR5, PtRR6, PtRR7, PtRR8, PtRR9, PtRR10, PtRR11 | Expression detected23; PtRR7 strongly downregulated in pBpCRE1:AtCKX2 plants32 |
| Zea maize ZmRR1/ZmCip1, ZmRR2, ZmRR3/ABPHYL1, ZmRR4, ZmRR5, ZmRR6, ZmRR7, ZmTcRR1, ZmTcRR2 | Upregulated by cytokinin (ZmRR1, ZmRR2)9,56,57, (ZmRR1,4–7)41; upregulated by nitrate in resupplied leaves (ZmRR1)8,9,57, (ZmRR2)55,57 localized in the nucleus (ZmRR3–7) or in the cytosol (ZmRR1–3)41; interaction of ZmRR1 with ZmHP1, ZmHP2 and ZmHP341; in vitro phosphorylation of ZmRR4 by ZmHP1 and ZmHP241; knockout of ZmRR3 shows decussate leaf pattern and a bigger meristem51–53; ZmTcRR1 and ZmTcRR2 are unusual response regulators expressed in the transfer cell layer33,54 |
| Vitis vinifera VvRRa1, VvRRa2, VvRRa3, VvRra4 | Regulated by cytokinin (VvRRa3 up and VvRRa4 down in isolated cells)7 |
Conclusions
In this review we summarized the state of the art in the field of cytokinin signaling beyond Arabidopsis. Some of the signaling components were identified in genetic screens while others were analyzed in systematic approaches following the sequencing of the respective genomes. Many studies link cytokinin to development, crosstalk with other hormones and also to other processes such as abiotic stress or nutrients deficiency response—just as has been shown for Arabidopsis. However, new aspects of cytokinin action that cannot be investigated in Arabidopsis were also discovered, clearly highlighting the necessity to look beyond a single model plant to understand the full spectra of cytokinin-regulated processes. Thus the benefits of cytokinin research in different plant species include: (i) examination of proteins or even whole protein families of the cytokinin signaling pathway that might behave differently than those in Arabidopsis—e.g., cytokinin inducible HPts and type-B RRs; (ii) indentifying the role of cytokinin in other pathways, morphological structures and developmental processes not present in Arabidopsis and (iii) using the wealth of information on the different signaling components to understand the evolution and the evolutionary trajectories of the cytokinin signaling pathway.
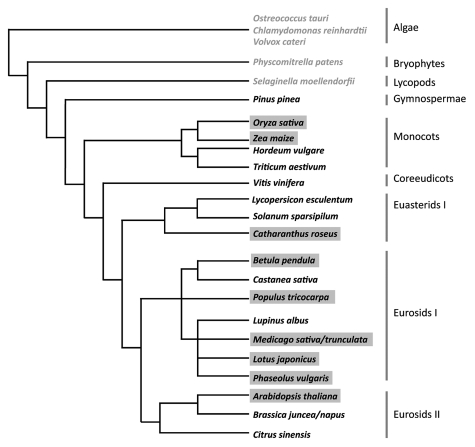
Currently experimental data concerning cytokinin signaling derive almost exclusively from angiosperm species (Fig. 1). A deeper insight into the evolution of this pathway requires the inclusion of more early-diverging plant species into this analysis. The rapidly increasing number of sequenced genomes combined with the already established tools of cytokinin research will enhance our understanding of this fascinating plant hormone. The future of cytokinin research is very bright, indeed.
Figure 1.
Phylogenetic relation of plant species used in cytokinin signaling research. Species written in grey were used in bioinformatic analysis.5 Black font marks species in which experimental data for cytokinin signaling components have been obtained on the RNA level. In species shaded in grey functional assays have been performed for members of the cytokinin signaling pathway (tree based on Sitte et al. 2002).65
Acknowledgements
This work was supported by a Elsa Neumann stipend to E.H., by a stipend from the Volkswagen Foundation to N.G. and support of the Dahlem Centre of Plant Science to A.H.. We are also very grateful to Anahid E. Powell for critically reading the manuscript.
Abbreviations
- TCS
two-component system
- HK
histidine kinase
- HPt
histidine phosphotransfer protein
- RR
response regulator
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13157
Supplementary Material
References
- 1.Santner A, Calderon-Villalobos LI, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- 2.Mok DW, Mok MC. Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- 3.Heyl A, Schmülling T. Cytokinin signal perception and transduction. Curr Opin Plant Biol. 2003;6:480–488. doi: 10.1016/s1369-5266(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 4.Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 5.Pils B, Heyl A. Unraveling the evolution of cytokinin signaling. Plant Physiol. 2009;151:782–791. doi: 10.1104/pp.109.139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deji A, Sakakibara H, Okumura S, Matsuda T, Ishida Y, Yamada S, et al. Accumulation of maize response regulator proteins in mesophyll cells after cytokinin treatment. Biosci Biotechnol Biochem. 2002;66:1853–1858. doi: 10.1271/bbb.66.1853. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes J, Tavares S, Amâncio S. Identification and expression of cytokinin signaling and meristem identity genes in sulfur deficient grapevine (Vitis vinifera L.) Plant Signal Behav. 2009;4:1128–1135. doi: 10.4161/psb.4.12.9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takei K, Sakakibara H, Taniguchi M, Sugiyama T. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 2001;42:85–93. doi: 10.1093/pcp/pce009. [DOI] [PubMed] [Google Scholar]
- 9.Takei K, Takahashi T, Sugiyama T, Yamaya T, Sakakibara H. Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J Exp Bot. 2002;53:971–977. doi: 10.1093/jexbot/53.370.971. [DOI] [PubMed] [Google Scholar]
- 10.Ding Y, Kalo P, Yendrek C, Sun J, Liang Y, Marsh JF, et al. Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell. 2008;20:2681–2695. doi: 10.1105/tpc.108.061739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Rizzo S, Crespi M, Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- 13.Papon N, Vansiri A, Gantet P, Chénieux JC, Rideau M, Crèche J. Histidine-containing phosphotransfer domain extinction by RNA interference turns off a cytokinin signalling circuitry in Catharanthus roseus suspension cells. FEBS Lett. 2004;558:85–88. doi: 10.1016/S0014-5793(03)01522-9. [DOI] [PubMed] [Google Scholar]
- 14.Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, et al. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- 15.Jolivet K, Grenier E, Bouchet JP, Esquibet M, Kerlan MC, Caromel B, et al. Identification of plant genes regulated in resistant potato Solanum sparsipilum during the early stages of infection by Globodera pallida. Genome. 2007;50:422–427. doi: 10.1139/g07-015. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava S, Srivastava AK, Suprasanna P, D'Souza SF. Comparative biochemical and transcriptional profiling of two contrasting varieties of Brassica juncea L. in response to arsenic exposure reveals mechanisms of stress perception and tolerance. J Exp Bot. 2009;60:3419–3431. doi: 10.1093/jxb/erp181. [DOI] [PubMed] [Google Scholar]
- 17.Jain M, Tyagi AK, Khurana JP. Differential gene expression of rice two-component signaling elements during reproductive development and regulation by abiotic stress. Funct Integr Genomics. 2008;8:175–180. doi: 10.1007/s10142-007-0063-6. [DOI] [PubMed] [Google Scholar]
- 18.Karan R, Singla-Pareek SL, Pareek A. Histidine kinase and response regulator genes as they relate to salinity tolerance in rice. Funct Integr Genomics. 2009;9:411–417. doi: 10.1007/s10142-009-0119-x. [DOI] [PubMed] [Google Scholar]
- 19.Du L, Jiao F, Chu J, Jin G, Chen M, Wu P. The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics. 2007;89:697–707. doi: 10.1016/j.ygeno.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Ishida K, Niwa Y, Yamashino T, Mizuno T. A genome-wide compilation of the two-component systems in Lotus japonicus. DNA Res. 2009;16:237–247. doi: 10.1093/dnares/dsp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito Y, Kurata N. Identification and characterization of cytokinin-signalling gene families in rice. Gene. 2006;382:57–65. doi: 10.1016/j.gene.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Jain M, Tyagi AK, Khurana JP. Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa) BMC Plant Biol. 2006;6:1. doi: 10.1186/1471-2229-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramírez-Carvajal GA, Morse AM, Davis JM. Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol. 2008;177:77–89. doi: 10.1111/j.1469-8137.2007.02240.x. [DOI] [PubMed] [Google Scholar]
- 24.Ramírez-Carvajal GA, Morse AM, Dervinis C, Davis JM. The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol. 2009;150:759–771. doi: 10.1104/pp.109.137505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishopp A, Help H, Helariutta Y. Cytokinin signaling during root development. Int Rev Cell Mol Biol. 2009;276:1–48. doi: 10.1016/S1937-6448(09)76001-0. [DOI] [PubMed] [Google Scholar]
- 26.Heyl A, Werner T, Schmülling T. Cytokinin metabolism and signal transduction. In: Hedden P, Thomas S, editors. Plant hormone signaling, Annual Plant Reviews. Blackwell Publishing Ltd; 2006. pp. 93–123. [Google Scholar]
- 27.Perilli S, Moubayidin L, Sabatini S. The molecular basis of cytokinin function. Curr Opin Plant Biol. 2010;13:21–26. doi: 10.1016/j.pbi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 28.Coba de la Peña T, Cárcamo CB, Almonacid L, Zaballos A, Lucas MM, Balomenos D, et al. A salt stress-responsive cytokinin receptor homologue isolated from Medicago sativa nodules. Planta. 2008;227:769–779. doi: 10.1007/s00425-007-0655-3. [DOI] [PubMed] [Google Scholar]
- 29.Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, et al. Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 2006;140:221–234. doi: 10.1104/pp.105.070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merchan F, de Lorenzo L, Rizzo SG, Niebel A, Manyani H, Frugier F, et al. Identification of regulatory pathways involved in the reacquisition of root growth after salt stress in Medicago truncatula. Plant J. 2007;51:1–17. doi: 10.1111/j.1365-313X.2007.03117.x. [DOI] [PubMed] [Google Scholar]
- 31.Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Tormakangas K, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 32.Nieminen K, Immanen J, Laxell M, Kauppinen L, Tarkowski P, Dolezal K, et al. Cytokinin signaling regulates cambial development in poplar. Proc Natl Acad Sci USA. 2008;105:20032–20037. doi: 10.1073/pnas.0805617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muñiz LM, Royo J, Gómez E, Baudot G, Paul W, Hueros G. Atypical response regulators expressed in the maize endosperm transfer cells link canonical two component systems and seed biology. BMC Plant Biol. 2010;10:84. doi: 10.1186/1471-2229-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol. 2004;134:1654–1661. doi: 10.1104/pp.103.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romanov GA, Spíchal L, Lomin SN, Strnad M, Schmülling T. A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Anal Biochem. 2005;347:129–134. doi: 10.1016/j.ab.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 36.Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov GA, Strnad M, et al. Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 2004;45:1299–1305. doi: 10.1093/pcp/pch132. [DOI] [PubMed] [Google Scholar]
- 37.Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim RA, et al. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 2008;49:1429–1450. doi: 10.1093/pcp/pcn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papon N, Clastre M, Andreu F, Gantet P, Rideau M, Crèche J. Expression analysis in plant and cell suspensions of CrCKR1, a cDNA encoding a histidine kinase receptor homologue in Catharanthus roseus (L.) G Don J Exp Bot. 2002;53:1989–1990. doi: 10.1093/jxb/erf048. [DOI] [PubMed] [Google Scholar]
- 39.Coba de la Peña T, Cárcamo CB, Almonacid L, Zaballos A, Lucas MM, Balomenos D, et al. A cytokinin receptor homologue is induced during root nodule organogenesis and senescence in Lupinus albus L. Plant Physiol Biochem. 2008;46:219–225. doi: 10.1016/j.plaphy.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 40.Kumar V, Mills DJ, Anderson JD, Mattoo AK. An alternative agriculture system is defined by a distinct expression profile of select gene transcripts and proteins. Proc Natl Acad Sci USA. 2004;101:10535–10540. doi: 10.1073/pnas.0403496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asakura Y, Hagino T, Ohta Y, Aoki K, Yonekura-Sakakibara K, Deji A, et al. Molecular characterization of His-Asp phosphorelay signaling factors in maize leaves: implications of the signal divergence by cytokinin-inducible response regulators in the cytosol and the nuclei. Plant Mol Biol. 2003;52:331–341. doi: 10.1023/a:1023971315108. [DOI] [PubMed] [Google Scholar]
- 42.Sugawara H, Kawano Y, Hatakeyama T, Yamaya T, Kamiya N, Sakakibara H. Crystal structure of the histidine-containing phosphotransfer protein ZmHP2 from maize. Protein Sci. 2005;14:202–208. doi: 10.1110/ps.041076905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugawara H, Yamaya T, Sakakibara H. Crystallization and preliminary X-ray diffraction study of the histidine-containing phosphotransfer protein ZmHP1 from maize. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:366–368. doi: 10.1107/S1744309105006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Lorenzo L, Merchan F, Blanchet S, Megias M, Frugier F, Crespi M, et al. Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes. Plant Physiol. 2007;145:1521–1532. doi: 10.1104/pp.107.106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought and salt stress in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maul P, Bausher M, McCollum G, Mozoruk J, Niedz R. CsHPt1, a putative histidine containing phosphotransmitter protein induced during early somatic embryogenensis in Valencia sweet orange. Plant Science. 2006;170:44–53. [Google Scholar]
- 47.Chefdor F, Bénédetti H, Depierreux C, Delmotte F, Morabito D, Carpin S. Osmotic stress sensing in Populus: components identification of a phosphorelay system. FEBS Lett. 2005;580:77–81. doi: 10.1016/j.febslet.2005.11.051. [DOI] [PubMed] [Google Scholar]
- 48.Ma QH, Tian B. Characterization of a wheat histidine-containing phosphotransfer protein (HP) that is regulated by cytokinin-mediated inhibition of leaf senescence. Plant Sci. 2005;168:1507–1514. [Google Scholar]
- 49.Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 2005;44:314–333. doi: 10.1111/j.1365-313X.2005.02530.x. [DOI] [PubMed] [Google Scholar]
- 50.Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T. Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp Phosphorelay circuitry. Plant Cell Physiol. 2005;46:339–355. doi: 10.1093/pcp/pci033. [DOI] [PubMed] [Google Scholar]
- 51.Jackson D, Hake S. Control of phyllotaxy in maize by the abphyl1 gene. Development. 1999;126:315–323. doi: 10.1242/dev.126.2.315. [DOI] [PubMed] [Google Scholar]
- 52.Giulini A, Wang J, Jackson D. Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature. 2004;430:1031–1034. doi: 10.1038/nature02778. [DOI] [PubMed] [Google Scholar]
- 53.Lee BH, Johnston R, Yang Y, Gallavotti A, Kojima M, Travencolo BA, et al. Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 2009;150:205–216. doi: 10.1104/pp.109.137034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muñiz LM, Royo J, Gómez E, Barrero C, Bergareche D, Hueros G. The maize transfer cell-specific type-A response regulator ZmTCRR-1 appears to be involved in intercellular signalling. Plant J. 2006;48:17–27. doi: 10.1111/j.1365-313X.2006.02848.x. [DOI] [PubMed] [Google Scholar]
- 55.Deji A, Sakakibara H, Ishida Y, Yamada S, Komari T, Kubo T, et al. Genomic organization and transcriptional regulation of maize ZmRR1 and ZmRR2 encoding cytokinin-inducible response regulators. Biochim Biophys Acta. 2000;1492:216–220. doi: 10.1016/s0167-4781(00)00076-2. [DOI] [PubMed] [Google Scholar]
- 56.Sakakibara H, Hayakawa A, Deji A, Gawronski SW, Sugiyama T. His-Asp phosphotransfer possibly involved in the nitrogen signal transduction mediated by cytokinin in maize: molecular cloning of cDNAs for two-component regulatory factors and demonstration of phosphotransfer activity in vitro. Plant Mol Biol. 1999;41:563–573. doi: 10.1023/a:1006391304881. [DOI] [PubMed] [Google Scholar]
- 57.Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T. A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J. 1998;14:337–344. doi: 10.1046/j.1365-313x.1998.00134.x. [DOI] [PubMed] [Google Scholar]
- 58.Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T. Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 1998;429:259–262. doi: 10.1016/s0014-5793(98)00611-5. [DOI] [PubMed] [Google Scholar]
- 59.Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 2007;48:523–539. doi: 10.1093/pcp/pcm022. [DOI] [PubMed] [Google Scholar]
- 60.Cheng X, Jiang H, Zhang J, Qian Y, Zhu S, Cheng B. Overexpression of type-A rice response regulators, OsRR3 and OsRR5, results in lower sensitivity to cytokinins. Genet Mol Res. 2010;9:348–359. doi: 10.4238/vol9-1gmr739. [DOI] [PubMed] [Google Scholar]
- 61.Cortizo M, Alvarez JM, Rodríguez A, Fernández B, Ordás RJ. Cloning and characterization of a type-A response regulator differentially expressed during adventitious shoot formation in Pinus pinea L. J Plant Physiol. 2010;167:1023–1026. doi: 10.1016/j.jplph.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 62.Camacho Y, Martinez-Castilla L, Fragoso S, Vázquez S, Martínez-Barajas E, Coello P. Characterization of a type A response regulator in the common bean (Phaseolus vulgaris) in response to phosphate starvation. Physiol Plant. 2008;132:272–282. doi: 10.1111/j.1399-3054.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- 63.Papon N, Oudin A, Vansiri A, Rideau M, Chénieux JC, Créche J. Differential expression of two type-A response regulators in plants and cell cultures of Catharanthus roseus (L.) G Don J Exp Bot. 2003;54:1793–1795. doi: 10.1093/jxb/erg185. [DOI] [PubMed] [Google Scholar]
- 64.Papon N, Clastre M, Gantet P, Rideau M, Chénieux °C, Créche J. Inhibition of the plant cytokinin transduction pathway by bacterial histidine kinase inhibitors in Catharanthus roseus cell cultures. FEBS Lett. 2003;537:101–105. doi: 10.1016/s0014-5793(03)00102-9. [DOI] [PubMed] [Google Scholar]
- 65.Sitte P, Weiler EW, Kadereit JW, Bresinsky A, Körner C. Strasburger, Lehrbuch der Botanik. Berlin: Spektrum Akademischer Verlag; 2002. [Google Scholar]
- 66.Hren M, Nikolic P, Rotter A, Blejec A, Terrier N, Ravnikar M, Dermastia M, Gruden K. ‘Bois noir’ phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genomics. 2009;10:460. doi: 10.1186/1471-2164-10-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imin N, Goffard N, Mizamidin M, Rolfe BG. Genome-wide transcriptional analysis of super-embryogenic Medicago truncatula explant cultures. BMC Plant Biol. 2008;8:110. doi: 10.1186/1471-2229-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.